Extended Data Fig. 7: The binding specificity of NP220 to unintegrated HIV-1 DNA.
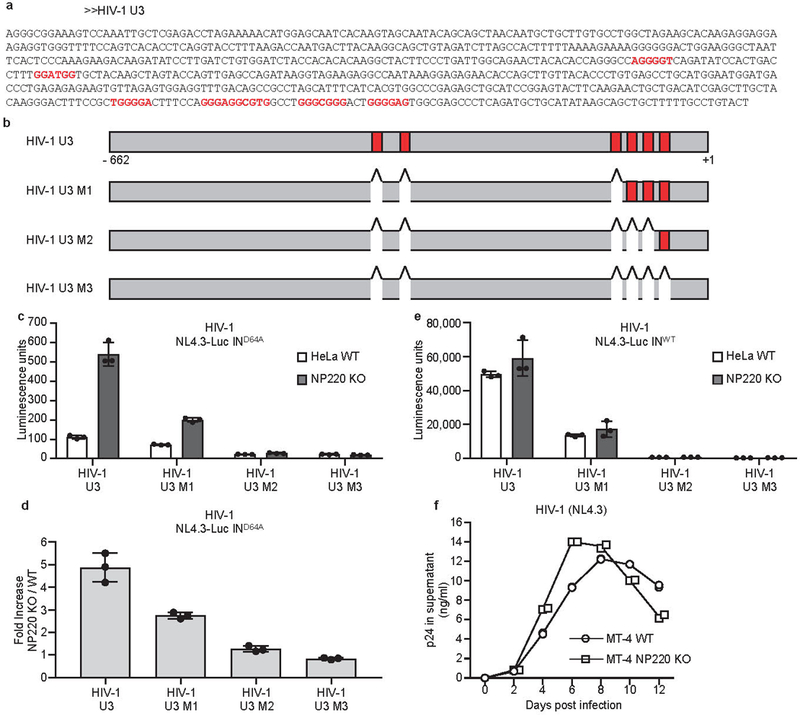
a, b, The sequences and locations of putative NP220 binding sites in HIV-1 U3 region of HIV-1 U3 region. Putative NP220 binding sites are indicated in red. c, d, Parental (WT) or NP220 knockout (NP220 KO) HeLa cells were infected with VSV-G pseudotyped, integrase-deficient (IND64A) MLV-Luc virus bearing indicated deletions or mutations in the U3 region. (c) Luciferase activities were measured 40 h post infection. (d) Fold increase (NP220 KO / WT) was calculated as the ratio of luciferase activities from KO cells to luciferase activities from WT cells. Data presented are mean ± SD from three independent experiments (n = 3).e, Parental (WT) or NP220 knockout (NP220 KO) HeLa cells were infected with VSV-G pseudotyped, integrase-proficient (INWT) HIV-Luc virus bearing indicated deletions or mutations in the U3 region. Luciferase activities were measured 40 h post infection. Data presented are mean ± SD from three independent experiments (n = 3). f, Knockout of NP220 increases the rate of HIV-1 spreading. (g) Parental (WT) and NP220 knockout (NP220 KO) MT-4 cells were infected with HIV-1(NL4.3). Viral spreading was monitored by assay for p24 concentration in the culture medium. Data presented are mean from two technical ELISA replicates and are representative of two independent experiments.
