Fig. 4: NP220 recruits HDAC1 and HDAC4 to deacetylate histone H3 on unintegrated retroviral DNA.
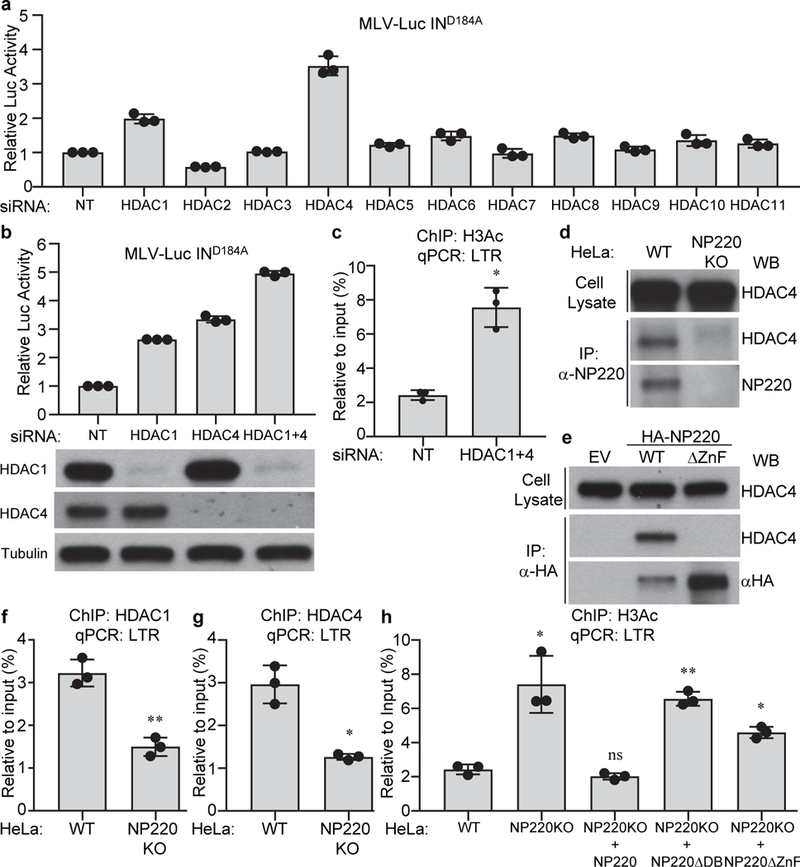
a-c, HDAC1/4 are required for the silencing of unintegrated retroviral DNA. Hela cells were transfected with indicated siRNAs and then infected with integrase-deficient (IND184A) MLV-Luc virus. (a, b) Luciferase activities were measured and luciferase activity in non-targeting (NT) siRNA transfected cells was set as 1. The expression of HDAC1 and HDAC4 was determined by Western blot (b, bottom panels). (c) ChIP was performed using antibodies to pan-acetyl H3 followed by qPCR targeting LTR. d, e, NP220 interacts with HDAC4. (d) Endogenous NP220 was immunoprecipitated (IP) from indicated HeLa cell lines. (e) HA-tagged NP220 or NP220 with Zinc finger deletion (ΔZnF) were introduced into NP220 KO HeLa cells, and then immunoprecipitated (IP) by HA antibody. The coimmunoprecipitating HDAC4 was probed by Western blot. Images are representative of two independent experiments with similar results. f-h, NP220 recruits HDAC1 and HDAC4 to deacetylate histone H3 on unintegrated retroviral DNA. Indicated cells were infected with integrase-deficient (IND184A) MLV-Luc virus. ChIP was performed using antibodies to HDAC1 (f), HDAC4 (g), pan-acetyl H3 (h), followed by qPCR targeting LTR. In (a-c and f-h), data presented are mean ± SD from three independent experiments (n = 3). ns: p > 0.05; *: p < 0.05; **: p < 0.01. P values are from paired two-sided t-tests. Exact p values are presented in the accompanying source data.
