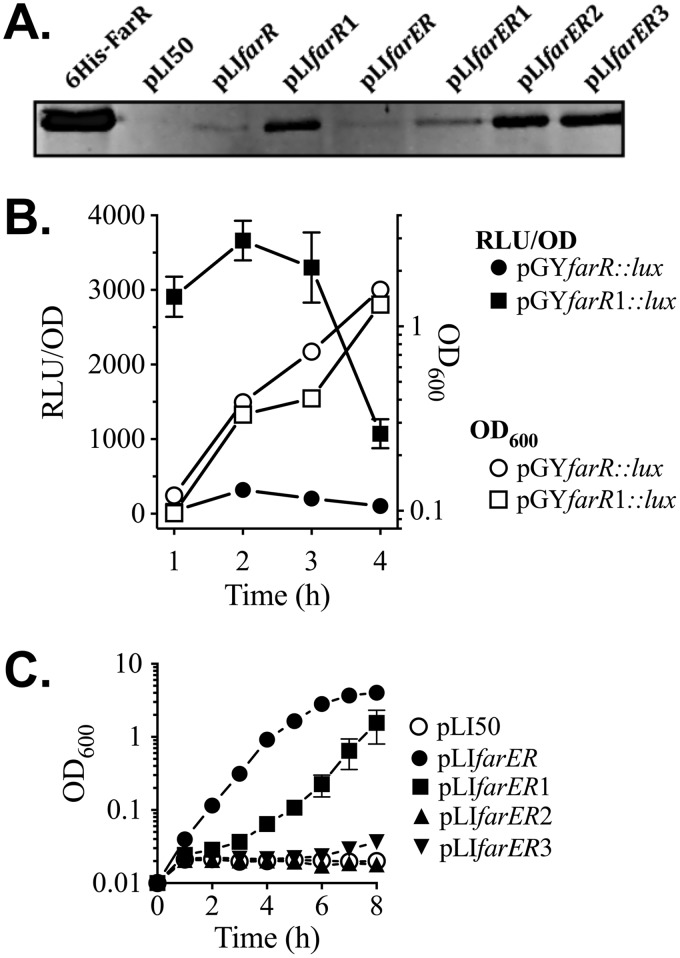
FIG 5.
Influence of nucleotide substitutions in −10PfarR and PAL1 on expression of FarR, and resistance to linoleic acid. (A) Western blot of gel loaded with 25 ng of purified 6×His-FarR or 25 µg of cell lysate protein from E. coli DH5α transformed with pLI50 vehicle, pLIfarR, or pLIfarER and derivatives containing nucleotide substitutions in −10PfarR (pLIfarR1 and pLIfarER1), PAL1 (pLIfarER2), or −10PfarR and PAL1 (pLIfarER3). (B) Luciferase reporter gene assays of S. aureus USA300 transformed with pGYfarR::lux, or pGYfarR1::lux harboring a G→A substitution in −10PfarR. Triplicates of each culture were grown in TSB, with monitoring of OD600 and luciferase activity (relative light units [RLU]) at hourly intervals. Data for RLU determinations are standardized to OD600 units (RLU/OD) to account for differences in cell density. (C) Growth of USA300 ΔfarER complemented with pLI50 vehicle or pLIfarER and derivatives in TSB plus 50 µM linoleic acid. Each data point represents the mean and standard deviation of the results from triplicate cultures.