Figure 4.
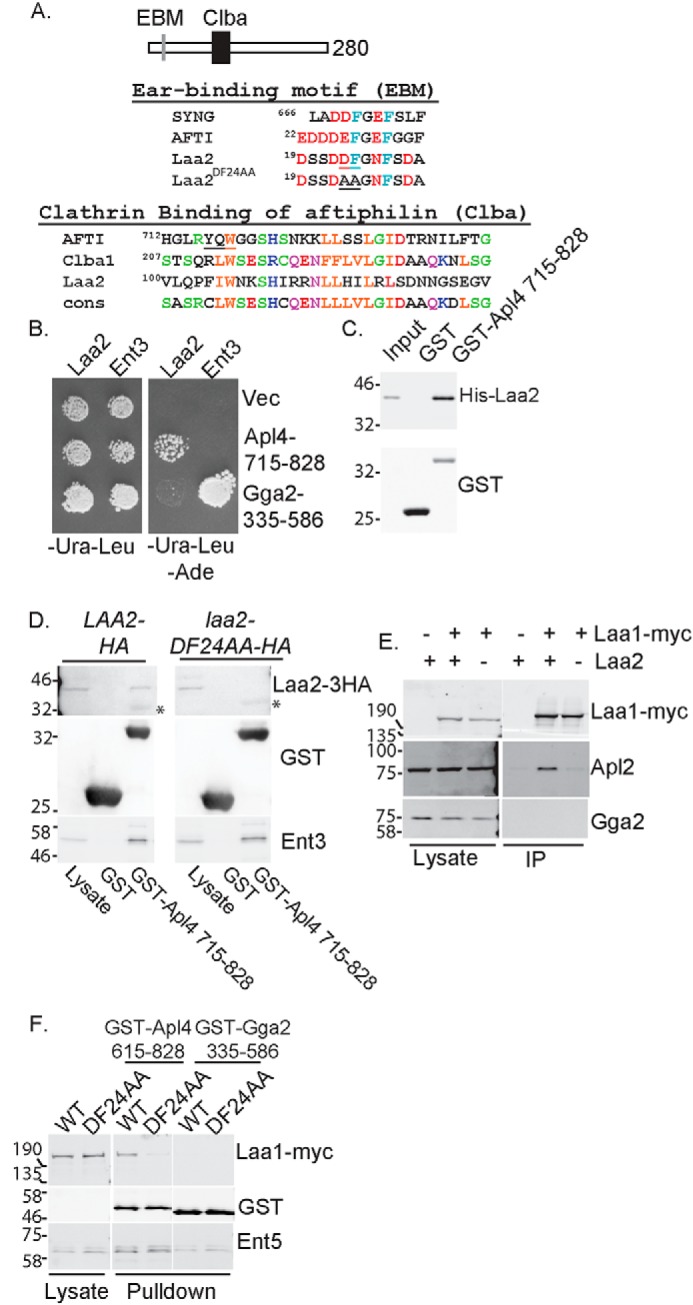
Laa2 ear-binding motif mediates the interaction with AP-1 γ-ear. A, schematic of Laa2 (top). Alignment of ear-binding motif (EBM) with those of γ-synergin and aftiphilin is shown. Underlined residues were mutated to alanines to generate laa2–DF24AA (middle). Alignment of Laa2 with Clba domains of aftiphilin, uncharacterized human protein Clba1, and the consensus motif for the domain defined in Pfam is shown. Underlined residues in aftiphilin are required for clathrin binding (bottom). B, Laa2 interacts with the γ-ear of AP-1 but not that of Gga2 in two-hybrid analysis. Cell expressing activation domain fusions to full-length Laa2 or Ent3, as a positive control, and DNA-binding domain fusions to indicated regions of Apl4 or Gga2 or empty vector were spotted on medium that selected for the plasmids (left) or medium that selected for interaction (right). C, Laa2 interacts directly with the γ-ear of AP-1. Recombinant Laa2 was subjected to GST pulldown using recombinant γ-ear of AP-1 or GST. Input is 10%. The data are representative of three repeats. D, Laa2 ear-binding motif is required for interaction with the γ-ear of AP-1 in GST pulldown experiments. Asterisks denote nonspecific binding of antibodies to GST. E, Laa2 is required for Laa1 interaction with AP-1. Laa1–Myc was immunoprecipitated from WT or laa2Δ cells. Endogenous AP-1 or Gga2 were detected with polyclonal antibodies. Lysates are 4% of immunoprecipitations. Data shown are representative of at least three repeats. F, Laa2 binding to AP-1 is required for Laa1 interaction with the γ-ear of AP-1. Cells expressing indicated allele of Laa2 and Laa1 from their endogenous loci were subjected to GST pulldown using the γ-ear of AP-1, or the γ-ear of Gga2, or GST as a negative control. Samples were probed for GST, HA, Myc, and the adaptor-interacting proteins Ent5 or Ent3, as a positive control. Lysates are 5 and 6% of pulldown without any GST protein for D and E, respectively. Locations of molecular weight standards are indicated. The data shown are representative of at least three repeats.
