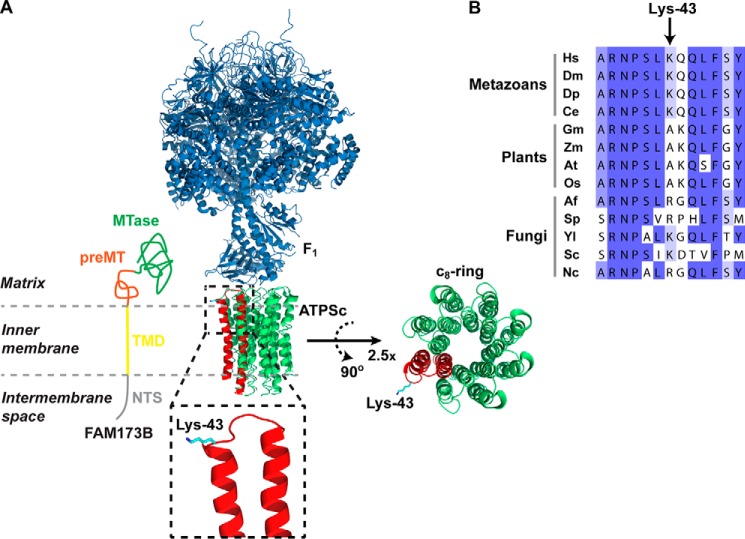
Figure 6.
Localization of Lys-43 in the ATPS complex structure and in the ATPSc sequence. A, structural representation of the complex between the F1 and ATPSc moieties (c8-ring) of the bovine ATPS complex, generated from a previously published structure (31) (Protein Data Bank code 2XND) and its topology in the inner mitochondrial membrane. Rotation and magnification (×2.5) of the c8-ring was performed to better visualize the ring structure. F1 is shown in blue, and ATPSc is shown in green, except for one of the eight protomers of the c8-ring, which is shown in red, and Lys-43 is here indicated in cyan. Also, a model for the submitochondrial localization of FAM173B is presented, with annotations and coloring as in Fig. 1. B, alignment of ATPSc-derived sequences from various organisms. Shown is the sequence surrounding the methylation site, Lys-43, in human ATPSc, aligned with the corresponding sequences of ATPSc from various organisms: H. sapiens (Hs), D. melanogaster (Dm), D. pulex (Dp), C. elegans (Ce), Glycine max (Gm), Zea mays (Zm), Arabidopsis thaliana (At), Oryza sativa (Os), Aspergillus fumigatus (Af), Schizosaccharomyces pombe (Sp), Yarrowia lipolytica (Yl), Saccharomyces cerevisiae (Sc), and Neurospora crassa (Nc). Sequence accession numbers are given in Table S3.