Abstract
Gram-negative bacteria produce outer-membrane vesicles (OMVs) that package genetic elements, virulence factors, and cell-to-cell communication signaling compounds. Despite their importance in many disease-related processes, how these versatile structures are formed is incompletely understood. A self-produced secreted small molecule, the Pseudomonas quinolone signal (PQS), has been shown to initiate OMV formation in Pseudomonas aeruginosa by interacting with the outer membrane (OM) and inducing its curvature. Other bacterial species have also been shown to respond to PQS, supporting a common biophysical mechanism. Here, we conducted molecular dynamics simulations to elucidate the specific interactions between PQS and a model P. aeruginosa OM at the atomistic scale. We discovered two characteristic states of PQS interacting with the biologically relevant membrane, namely attachment to the membrane surface and insertion into the lipid A leaflet. The hydrogen bonds between PQS and the lipid A phosphates drove the PQS–membrane association. An analysis of PQS trajectory and molecular conformation revealed sequential events critical for spontaneous insertion, including probing, docking, folding, and insertion. Remarkably, PQS bent its hydrophobic side chain into a closed conformation to lower the energy barrier for penetration through the hydrophilic headgroup zone of the lipid A leaflet, which was confirmed by the potential of mean force (PMF) measurements. Attachment and insertion were simultaneously observed in the simulation with multiple PQS molecules. Our findings uncover a sequence of molecular interactions that drive PQS insertion into the bacterial OM and provide important insight into the biophysical mechanism of small molecule–induced OMV biogenesis.
Keywords: molecular dynamics, Pseudomonas aeruginosa (P. aeruginosa), lipid A, small molecule, outer membrane, hydrogen bond, quorum sensing, cell communication, membrane dynamics, outer membrane vesicle biogenesis
Introduction
Outer-membrane vesicles (OMVs)2 derived from the bacterial outer membrane (OM) are instrumental in bacterial interactions. They are spherical membranous structures that can package cargoes, such as virulence factors, proteins, DNA, RNA, and signaling molecules (1–3). In contrast to the well-known functions of OMVs in trafficking systems, the biogenesis of OMVs is still under investigation. Several mechanisms for OMV formation have been proposed (3, 4). One hypothesis builds on the accumulation of chemical compounds (e.g. misfolded proteins (5) or muramyl peptides (6)) in the periplasmic space. These fragments create turgor pressure on the OM and further stimulate vesicle formation. Another proposed mechanism focuses on reduced cross-linking between the OM and peptidoglycan layer. The lack of trans-envelope proteins untethers the OM from the cell wall (7, 8), and vesiculation occurs when the OM grows faster than the peptidoglycan layer (9). Emphasizing the membrane surface, a third explanation highlights the intercalation of molecules, such as antibiotics, phospholipids, and signaling molecules, into the outer leaflet of OM (4, 10, 11). Li et al. (10) discussed that the presence of cationic antibiotics destabilizes the OM by collapsing salt bridges between lipopolysaccharides (LPSs) and divalent ions. The disturbed OM then forms OMVs. Roier et al. (4) showed that disruption of the VacJ/Yrb ABC phospholipid transport system increases phospholipid number in the OM outer leaflet, causing OMVs enriched in phospholipids to eventually detach from the membrane surface.
We developed the bilayer-couple model (11) to describe OMV biogenesis in Pseudomonas aeruginosa from a purely biophysical perspective. This model is based on the stimulation of OMV production by interactions of a self-produced small molecule, 2-heptyl-3-hydroxy-4-quinolone (the Pseudomonas quinolone signal (PQS)), with the OM (12, 13). The overall amphiphilicity of PQS and its task-specific functional groups enable a strong association with LPS in the outer leaflet (13). The insertion of PQS into the membrane leads to an asymmetric expansion of the outer leaflet compared with the inner leaflet, which drives the induction of membrane curvature and subsequent pinch-off of the OMV (11, 14). Recent experiments have shown that other gammaproteobacteria also respond to PQS by increasing OMV formation, which suggests that small molecule–induced OMV biogenesis is a general mechanism that is driven by biophysical interactions with target membranes (15). Previous experiments have suggested that strong interactions between PQS and the lipid A portion of LPS may be dictated by hydrophobic forces and hydrogen bonding (13). However, it is challenging to probe specific interactions and the corresponding conformation and orientation of PQS when interacting with surrounding lipids on the nanoscale while using only experimental techniques. In this work, we utilized molecular dynamics simulations to reveal atomistic details of the physical interactions between PQS and the P. aeruginosa OM and to discover the dynamic behavior of the PQS–membrane association.
Results
PQS has five featured functional groups as shown in Fig. 1. They are phenyl, amine, second-position alkyl chain, third-position hydroxyl, and fourth-position carbonyl. We conducted preliminary simulations to characterize the interaction between water and PQS as well as the aggregation behavior of multiple PQS molecules. The results show that the quinolone structure (“head”) of PQS is amphiphilic. In particular, carbonyl and hydroxyl are hydrophilic sites, whereas phenyl and amine are hydrophobic (see supporting information and Figs. S1 and S2 for a detailed discussion). The electrostatic potential surface in Fig. 1, inset, shows strong electronegativity of carbonyl and hydroxyl as well as electropositivity of heterocyclic amine. These three functional groups will participate in hydrogen bond formation when interacting with the lipid A phosphates as demonstrated in the simulation results below.
Figure 1.
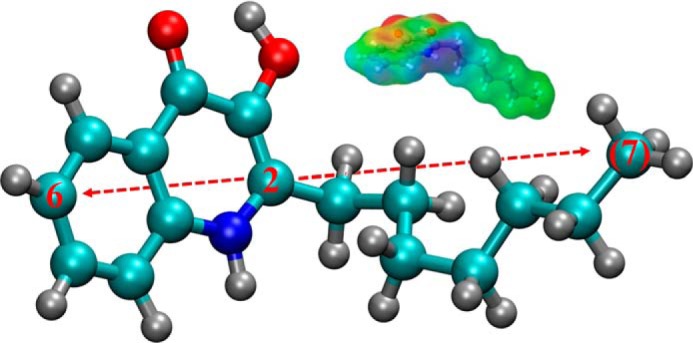
Chemical structure of PQS. Atoms are represented in different colors: cyan for carbon, silver for hydrogen, red for oxygen, and blue for nitrogen. Selected carbon atoms are numbered for describing the molecular conformation with the parentheses indicating the one in the alkyl side chain. The inset is the molecular electrostatic potential surface of PQS obtained by quantum mechanics calculation. The color map was chosen to represent negative potential in red and positive potential in blue.
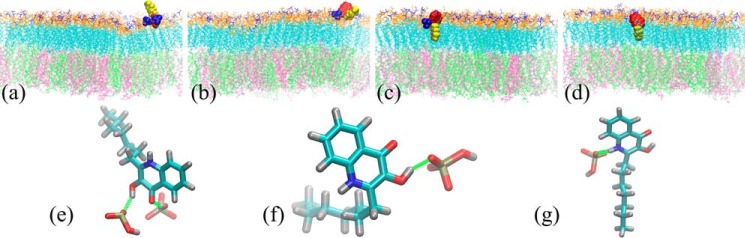
The independent unrestrained simulations were conducted by placing a single PQS molecule with different initial orientations in the aqueous phase in close proximity to the surface of lipid A leaflet. The representative snapshots of these simulations are shown in Fig. 2. During the first simulation, PQS first randomly moved in the aqueous phase above the membrane surface and attempted to establish interactions with the lipid A leaflet (“probing”). The small molecule then quickly attached to the membrane surface as shown in Fig. 2a. The head of PQS was embedded in the lipid A headgroups, whereas the hydrophobic alkyl side chain (“tail”) extended into the aqueous phase. The lipid A phosphates formed hydrogen bonds (H-bonds) with the third-position hydroxyl and fourth-position carbonyl of PQS (see Fig. 2e). This tail-up attachment was maintained in the equilibrium state (see Video S1 and the final snapshot in Fig. S3). The variations of the H-bond numbers and the corresponding pair number during the attachment are plotted in Fig. 3a and Fig. S4a, respectively.
Figure 2.
Snapshots of the first unrestrained simulation at 30 ns (a) and the second unrestrained simulation at 180 (b), 400 (c), and 500 ns (d) are shown. The zoom-in views of the hydrogen bonds between PQS and the associated lipid A phosphates (marked as green dashed lines for clarity) in a–c are presented in e–g. The heterocyclic rings and heptyl group in PQS are, respectively, colored in red and yellow in a–d. Functional groups in lipid A are represented in different colors: blue for phosphate, orange for 3-(acetyl amino)-3-deoxy-d-glucose, and cyan for dodecanoic acid (12:0) and 3-hydroxydecanoic acid 10:0 (3-OH). 1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, and 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol molecules are colored in silver, lime, and mauve, respectively. The color scheme in e–g is the same as Fig. 1 with the phosphorus atom in tan.
Figure 3.
The time evolution of hydrogen bond number (a and d), minimum distance between PQS and phospholipid leaflet (b and e), and head–tail angle of PQS (c and f). The top and bottom panels are, respectively, the results of the first and second unrestrained simulations. The criterion for hydrogen bond formation is a donor–acceptor distance of 0.35 nm and an angle of 30° (16).
Contributions from different functional groups in PQS were identified during the interaction. We found that the stable attachment formed after 200 ns was primarily attributed to the hydroxyl H-bonds, whereas the carbonyl H-bonds contributed mainly to the initial contact within the first 100 ns. The H-bonds between the PQS amine and the lipid A phosphates were observed, but their presence was sporadic.
The second unrestrained simulation showed more intriguing dynamics. Similarly, PQS probed the membrane surface under thermal fluctuations in the first 100 ns. The engagement of hydroxyl and carbonyl H-bonds facilitated the early anchoring of PQS at the lipid A surface (Fig. 2, b and f) from 100 to 200 ns. However, PQS migrated substantially along the membrane surface with constantly varying orientation and conformation during this preliminary association, which we refer to as the “docking” stage (see Video S2). We observed spontaneous insertion of PQS into the lipid A leaflet around 200 ns, which was exclusively associated with persistent amine H-bonds (Fig. 2, c and g). Notably, PQS moved slightly deeper into the lipid A leaflet during the last 100 ns of the simulation as depicted in Fig. 2d. The corresponding vertical displacement of PQS was about 0.2 nm. Fig. 3d shows that no H-bonds were detected afterward because of the increased donor–acceptor distance, which substantially exceeded the criterion of 0.35 nm (see the H-bond pair number in Fig. S4b) (16). This dynamic process indicates that the amine plays a critical role in the spontaneous insertion of PQS by forming temporary H-bonds with nearby phosphates. After the insertion, strong hydrophobic forces between the lipid A acyl chains and the PQS alkyl group drive PQS to penetrate deeper to promote the contact between long hydrocarbon chains. This displacement leads to the breakage of amine H-bonds.
We further probed the dynamics of attachment and insertion of PQS in detail. The insertion depth of PQS was measured by the smallest distance between PQS atoms and inner leaflet atoms. The conformation of PQS was characterized by the head–tail angle, which is the angle between the vector pointing from carbon 2 to carbon 6 and that pointing from carbon 2 to carbon (7)3 of the heptyl group as indicated by the arrows in Fig. 1. Fig. 3, b and c, show two stages of the PQS–membrane interaction in the first simulation, namely probing and attachment. The minimum distance obtained was constant, highlighting the stability of attachment. The large head–tail angle indicates that PQS adopted an open conformation. In contrast, the second simulation (Fig. 3, e and f) exhibited a four-stage dynamic process. In particular, PQS moved randomly in the aqueous phase from the start of simulation to 105 ns. The minimum distance took a relatively low value between 105 and 170 ns but with sporadic peaks, indicating unstable docking of PQS. More importantly, we discovered a subsequent folding stage in the 170–198-ns interval, during which the minimum distance further decreased and the head–tail angle suddenly dropped ∼40°. In other words, PQS bent its alkyl chain to adopt a closed, V-shaped conformation to promote its penetration through the lipid A headgroup zone. The insertion eventually occurred around 198 ns as PQS straightened the alkyl chain to align with the lipid A moiety.
The attachment stage in the first simulation and the docking stage in the second simulation exhibited similar behavior. In general, PQS was ∼1.35 nm above the phospholipid layer. Additionally, the H-bonds between PQS and the lipid A phosphates were dominated by the hydroxyl group. The thrusting of PQS into the lipid A layer can be directly perceived from the abrupt decrease in the minimum distance near 208 ns marked by the dashed red line in Fig. 3e. The folding of the alkyl chain took place right before the insertion. These results imply that the conformational transition is critical for the spontaneous insertion of PQS into the lipid A membrane leaflet.
The potential of mean force (PMF) analysis was conducted to provide quantitative evidence of the conformational dependence of the PQS–lipid A interaction. Two characteristic PMF profiles (Fig. 4a) were obtained from two transmembrane pulling simulations with different PQS entering poses (Fig. 4, b and c), namely the closed conformation (Simulation 1, the red line) and the open conformation with the tail-up orientation (Simulation 2, the blue line). These two conformations closely resembled the characteristic conformations observed in the unrestrained simulations. The slope of the PMF curve around the center-of-mass distance of −2 reflects the total free energy change of the system when PQS penetrated the lipid A headgroup zone. The slope is clearly smaller for the simulation with PQS in the closed conformation when it was pulled through the lipid A leaflet. This indicates that PQS reduced the repulsive forces from the lipid A headgroups during insertion by minimizing the exposure of hydrophobic alkyl chain, which was achieved by folding it against the rings. To follow up, we examined the 10-ns equilibrium of selected sampling windows in the PMF analysis. A conformational transition from the closed state to the open state was confirmed (see Fig. S5), which is consistent with the observation in the unrestrained simulations. The opening of alkyl chain allows its insertion into the hydrophobic region of lipid A. These intriguing conformational dynamics significantly reduce the repulsion between PQS and the membrane as shown in Fig. S5d.
Figure 4.

a, the potential of mean force for PQS as a function of the vertical distance from the membrane's center of mass (COM) (negative distance corresponds to a PQS position above the outer leaflet). The shaded green region roughly identifies the space occupied by the membrane. Two umbrella simulations with different initial orientations of PQS were performed. The red and blue lines represent the simulations with PQS orientation and conformation in contact with the membrane surface as in b and c, respectively.
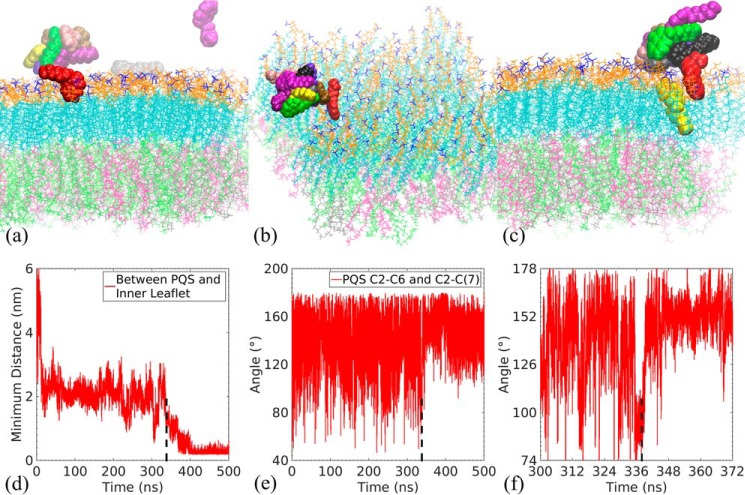
To further confirm the characteristic attachment and spontaneous insertion, we conducted unrestrained simulations with 10 PQS molecules initially dispersed in the solvent (Video S3). In other words, we significantly increased the PQS concentration in the aqueous phase. The snapshots and the association dynamics are presented in Fig. 5. Due to the interactions between the heterocyclic rings (see supporting information for a detailed discussion), PQS molecules were prone to aggregate in the solvent. However, assembly and disassembly of PQS aggregates was dynamic in the presence of a nearby membrane surface. We observed that one PQS quickly attached to the lipid A leaflet with the tail-up orientation around 42 ns (see Fig. 5a), resembling the stable attachment state observed in the single-molecule simulation. Other PQS molecules aggregated in the aqueous phase. The PQS cluster drifted along the membrane surface. Upon its interaction with the cluster, the attached PQS served as an anchor that localized the cluster as shown in Fig. 5b. At ∼338 ns in Fig. 5d, a PQS within the localized cluster spontaneously inserted into the membrane. Compared with the single PQS system, the insertion was delayed in the presence of neighboring PQS molecules as the self-interaction among PQSs flattened individual molecules and hindered their folding. In contrast to the 28-ns folding stage in Fig. 3f, which exhibited an ∼40° change in the head–tail angle, a shorter folding phase with a smaller angle change was observed in Fig. 5, e and f. Interestingly, we simultaneously observed both the attached and inserted states at the end of multiple-PQS simulation in Fig. 5c.
Figure 5.
Snapshots of 500-ns unrestrained simulations with 10 PQSs at 42 (a), 151 (b), and 500 ns (c) are shown. The time evolution of minimum distance between PQS and phospholipid leaflet (d), the head–tail angle of PQS (e), and enlarged view of the head–tail angle variations around the insertion instant of the 10-PQS system (f) are also shown.
Discussion
PQS is a self-produced small molecule known to stimulate OMV biogenesis in P. aeruginosa through a biophysical mechanism (11–13, 17). This paradigm of small molecule–induced OMV biogenesis could easily be generalized to other species, although no other candidate small molecule has yet been discovered. Other hydrophobic secreted molecules, such as cis-Δ2-11-methyl-dodecenoic acid in Stenotrophomonas maltophilia and cis-Δ2-dodecenoic acid in Burkholderia cenocepacia, have been connected to OMV formation in their respective species (18), but their effects have yet to be untangled from quorum-sensing signaling in the way they have been for PQS. Our recent work showed that exogenously added PQS induces OMV biogenesis in several Gram-negative species, and these other species also secrete OMV-inducing factors (15), strengthening the case for widespread use of a small molecule–initiated mechanism for OMV production. It is essential, therefore, that we characterize the interactions that curvature-inducing small molecules like PQS make with the outer membrane so we can understand how they drive this process. In the work presented here, we quantitatively assessed the nature and dynamics of PQS–lipid A interactions using our recently developed model of the P. aeruginosa OM (19) and made a number of intriguing discoveries. Our analyses highlighted a critical, and previously unknown, role for hydrogen bonding through the heterocyclic amine group in PQS that allows it to penetrate into the membrane. Much weight had previously been given to the 3-hydroxyl group due to experimental evidence demonstrating that 2-heptyl-4-quinolone, identical in structure to PQS but lacking the 3-hydroxyl group, was incapable of inducing either OMV formation in P. aeruginosa (20) or curvature in membrane surrogates (11). 2-Heptyl-4-quinolone also did not interact with LPS aggregates in the same way that PQS did (13). Our work here confirms the importance of the 3-hydroxyl group but suggests a different function from what was previously thought. Rather than forming long-term hydrogen bonds with lipid A in the OM, our analysis shows that this group primarily serves as an initiator of the PQS–OM interaction, docking the molecule in a favorable location to insert. Spontaneous insertion into the OM occurred only when the amine–phosphate hydrogen bonds were subsequently formed. Concomitant with PQS insertion into the membrane, the molecule also underwent a fascinating conformational folding that lowered the energy required to traverse the phosphate-rich region of the lipid A leaflet and facilitated engagement of the amine–phosphate hydrogen bond. We suspect that the sequence of events outlined here, docking–folding–insertion, may not be unique to the P. aeruginosa PQS system but may serve as a template for the interaction of OMV-inducing molecules with the OM of many Gram-negative organisms. We are currently investigating small-molecule and lipid derivatives to test this hypothesis and gain insight into the variable PQS responses across species that we noted in our experimental studies (15). Nevertheless, the discoveries in this work deliver unprecedented atomistic detail about the interactions of PQS with the OM, providing critical new insight while supporting previous physical experiments. Moreover, PQS dynamically remodels the associated membrane and can induce membrane curvature as proposed by the bilayer-couple model. Future work will also characterize the membrane curvature field induced by PQS insertion to test this hypothesis.
In conclusion, our computational study discovered a closed conformation of PQS in advance of its spontaneous insertion into the lipid A leaflet of the OM, which is confirmed universally in the single-molecule and multiple-PQS systems as well as in both unrestrained and PMF pulling simulations. The results demonstrate that the folding of the alkyl side chain is the critical step that decides whether PQS successfully inserts into the membrane or just attaches on the membrane surface. The findings suggest that the conformation of the small molecule significantly influences its interaction with the bacterial OM and could break new ground for answering questions about the biochemical and biophysical nature of OMV biogenesis across multiple species.
Experimental procedures
A model OM with rough LPS chemotype of P. aeruginosa was considered in this study. The outer leaflet was constructed with penta-acylated lipid A, and the inner leaflet was a mixture of phospholipids, reproducing the physical composition of P. aeruginosa OM (19). The asymmetric membrane was modeled by a hybrid GLYCAM-Slipids force field (FF) (1). The membrane construction is detailed in the supporting information, and the structural analysis of the membrane was reported previously (19). To be compatible with the membrane FF, the general AMBER FF (21) was selected to simulate PQS. The partial charges of PQS were derived by the restricted electrostatic potential approach (22) using the R.E.D. tool (23).
A PQS molecule was dispersed in water and equilibrated for 10 ns at 300 K and 1 bar before introduction to the membrane system. Unrestrained simulations of 300–500 ns were carried out at 323 K and 1 bar with PQS initially placed at 1 nm above the lipid A leaflet. Umbrella sampling simulations were conducted for calculating PMF profiles of PQS interacting with the model membrane. The initial configurations for the umbrella sampling were obtained by vertically pulling PQS through the membrane at a rate of 0.005 nm ps−1 with a force constant of 1000 kJ mol−1 nm−2. 79 sampling windows with a spacing of 0.2 nm were selected, and each sampling simulation was 10 ns. Total simulation time for obtaining one PMF was ∼790 ns. All simulations were performed using the GROMACS 5.1.2 package (24–29). Other computational details are described in the supporting information.
Author contributions
A. L. data curation; A. L. formal analysis; A. L. validation; A. L. and X. Y. investigation; A. L. and X. Y. methodology; A. L. writing-original draft; A. L., J. W. S., and X. Y. writing-review and editing; J. W. S. and X. Y. conceptualization; J. W. S. and X. Y. funding acquisition; J. W. S. and X. Y. project administration; X. Y. supervision.
Supplementary Material
Acknowledgments
Computing time was allocated by the Watson Data Center at Binghamton University and the Center for Computational Research at the University at Buffalo through the Virtual Infrastructure for Data Intensive Analysis (VIDIA).
This work was supported by NIAID, National Institutes of Health Grant R21AI121848. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5, Videos S1–S3, and supporting information.
In this study, the side chain carbons are numbered from (1) to (7), while the atoms in the main heterocyclic ring are numbered from 1 to 8.
- OMV
- outer-membrane vesicle
- PQS
- Pseudomonas quinolone signal (2-heptyl-3-hydroxy-4-quinolone)
- OM
- outer membrane
- PMF
- potential of mean force
- H-bond
- hydrogen bond
- FF
- force field
- LPS
- lipopolysaccharide.
References
- 1. Beveridge T. J. (1999) Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schertzer J. W., and Whiteley M. (2013) Bacterial outer membrane vesicles in trafficking, communication and the host-pathogen interaction. J. Mol. Microbiol. Biotechnol. 23, 118–130 10.1159/000346770 [DOI] [PubMed] [Google Scholar]
- 3. Vella B. D., and Schertzer J. W. (2015) Understanding and exploiting bacterial outer membrane vesicles, in Pseudomonas (Ramos J.L., Goldberg J. B., and Filloux A., eds) pp. 217–250, Springer, Dordrecht, The Netherlands: 10.1007/978-94-017-9555-5_9 [DOI] [Google Scholar]
- 4. Roier S., Zingl F. G., Cakar F., Durakovic S., Kohl P., Eichmann T. O., Klug L., Gadermaier B., Weinzerl K., Prassl R., Lass A., Daum G., Reidl J., Feldman M. F., and Schild S. (2016) A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7, 10515 10.1038/ncomms10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McBroom A. J., and Kuehn M. J. (2007) Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63, 545–558 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou L., Srisatjaluk R., Justus D. E., and Doyle R. J. (1998) On the origin of membrane vesicles in Gram-negative bacteria. FEMS Microbiol. Lett. 163, 223–228 10.1111/j.1574-6968.1998.tb13049.x [DOI] [PubMed] [Google Scholar]
- 7. Bernadac A., Gavioli M., Lazzaroni J. C., Raina S., and Lloubès R. (1998) Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180, 4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deatherage B. L., Lara J. C., Bergsbaken T., Rassoulian Barrett S. L., Lara S., and Cookson B. T. (2009) Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395–1407 10.1111/j.1365-2958.2009.06731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wensink J, and Witholt B. (1981) Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur. J. Biochem. 116, 331–335 10.1111/j.1432-1033.1981.tb05338.x [DOI] [PubMed] [Google Scholar]
- 10. Li Z., Clarke A. J., and Beveridge T. J. (1996) A major autolysin of Pseudomonas aeruginosa: subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J. Bacteriol. 178, 2479–2488 10.1128/jb.178.9.2479-2488.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schertzer J. W., and Whiteley M. (2012) A bilayer-couple model of bacterial outer membrane vesicle biogenesis. MBio 3, e00297-11 10.1128/mBio.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mashburn L. M., and Whiteley M. (2005) Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425 10.1038/nature03925 [DOI] [PubMed] [Google Scholar]
- 13. Mashburn-Warren L., Howe J., Garidel P., Richter W., Steiniger F., Roessle M., Brandenburg K., and Whiteley M. (2008) Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 69, 491–502 10.1111/j.1365-2958.2008.06302.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Florez C., Raab J. E., Cooke A. C., and Schertzer J. W. (2017) Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. MBio 8, e01034-17 10.1128/mBio.01034-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horspool A. M., and Schertzer J. W. (2018) Reciprocal cross-species induction of outer membrane vesicle biogenesis via secreted factors. Sci. Rep. 8, 9873 10.1038/s41598-018-28042-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Spoel D., van Maaren P. J., Larsson P., and Tîmneanu N. (2006) Thermodynamics of hydrogen bonding in hydrophilic and hydrophobic media. J. Phys. Chem. B 110, 4393–4398 10.1021/jp0572535 [DOI] [PubMed] [Google Scholar]
- 17. Schertzer J. W., Brown S. A., and Whiteley M. (2010) Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol. Microbiol. 77, 1527–1538 10.1111/j.1365-2958.2010.07303.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Devos S., Van Oudenhove L., Stremersch S., Van Putte W., De Rycke R., Van Driessche G., Vitse J., Raemdonck K., and Devreese B. (2015) The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front. Microbiol. 6, 298 10.3389/fmicb.2015.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li A., Schertzer J. W., and Yong X. (2018) Molecular dynamics modeling of Pseudomonas aeruginosa outer membranes. Phys. Chem. Chem. Phys. 20, 23635–23648 10.1039/C8CP04278K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mashburn-Warren L., Howe J., Brandenburg K., and Whiteley M. (2009) Structural requirements of the Pseudomonas quinolone signal for membrane vesicle stimulation. J. Bacteriol. 191, 3411–3414 10.1128/JB.00052-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., and Case D. A. (2004) Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- 22. Bayly C. I., Cieplak P., Cornell W., and Kollman P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 10.1021/j100142a004 [DOI] [Google Scholar]
- 23. Dupradeau F.-Y., Pigache A., Zaffran T., Savineau C., Lelong R., Grivel N., Lelong D., Rosanski W., and Cieplak P. (2010) The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 12, 7821–7839 10.1039/c0cp00111b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berendsen H. J. C., van der Spoel D., and van Drunen R. (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 10.1016/0010-4655(95)00042-E [DOI] [Google Scholar]
- 25. Lindahl E., Hess B., and van der Spoel D. (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7, 306–317 10.1007/s008940100045 [DOI] [Google Scholar]
- 26. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., and Berendsen H. J. (2005) GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- 27. Hess B., Kutzner C., van der Spoel D., and Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- 28. Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., and Lindahl E. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 10.1093/bioinformatics/btt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Páll S., Abraham M. J., Kutzner C., Hess B., and Lindahl E. (2015) Tackling exascale software challenges in molecular dynamics simulations with GROMACS, in Solving Software Challenges for Exascale. EASC 2014. Lecture Notes in Computer Science (Markidis S., and Laure E., eds) Vol. 8759, pp. 3–27, Springer, Cham, Switzerland: 10.1007/978-3-319-15976-8_1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.