Figure 1.
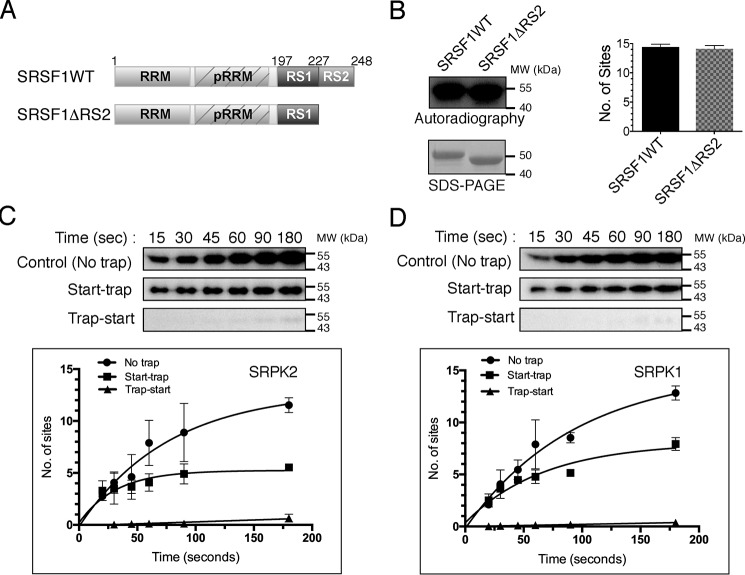
SRPK2 phosphorylates SRSF1 in a less processive manner. A, domain organization of SRSF1WT and SRSF1ΔRS2 (aa 1–226) are shown. pRRM, pseudo-RRM. B, phosphorylation of GST-tagged SRSF1WT (1 μm) and SRSF1ΔRS2 (1 μm) by SRPK2 (1.5 μm) in the presence of 100 μm [32P]ATP for 15 min was visualized by autoradiography (top left panel). SDS-PAGE analysis of the SRSF1 proteins used for the kinase assay (bottom left panel). The number of phosphorylation sites was quantitated by scintillation counting. SRSF1 contained 14.4 ± 0.3 phosphorylated sites, whereas SRSF1ΔRS2 contained 14.1 ± 0.4 sites (right panel). Phosphorylation of GST-tagged SRSF1 by SRPK2 (C) and SRPK1 (D) is shown under different trapping conditions. No trap (positive control), trap-start (negative control), and start-trap reactions are indicated in each case. In the start-trap experiment, SRPK1 or SRPK2 (1 μm) was pre-equilibrated with 300 nm SRSF1 and then allowed to react with 100 μm [32P]ATP in the absence (no trap) and the presence (start-trap) of 40 μm SRPK1KD or SRPK2KD (the corresponding kinase-dead mutant), respectively, added at the reaction start time. The kinase-dead mutant was added prior to the reaction start time in the negative control experiment (trap-start). SRSF1 was processively phosphorylated by SRPK2 to 5.2 ± 0.4 sites and SRPK1 to 7.5 ± 0.5 sites. Error bars, S.D. from three independent experiments.