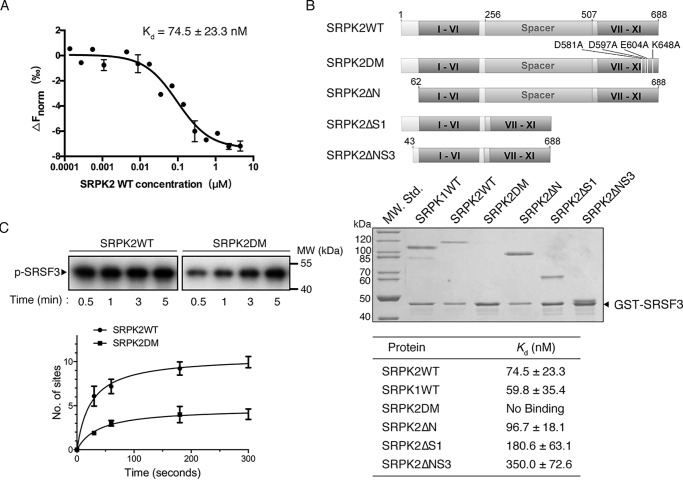
Figure 3.
Docking groove of SRPK2 is important for SRSF3 binding. A, binding affinity of SRSF3WT and SRPK2 analyzed by MicroScale thermophoresis. SRSF3WT was labeled and used at a concentration of 43.5 nm, whereas SRPK2 was titrated to concentrations between 4.5 μm and 0.137 nm. A Kd of 74.5 ± 23.3 nm was determined for this interaction (5-s MST-on time for evaluation). The graph displays data from three independent measurements (error bars represent the S.D.). B, domain organizations of SRPK2 and the different constructs were generated to delineate the region responsible for interaction with GST-SRSF3 (top). GST pulldown assay samples were separated by SDS-PAGE followed by Coomassie Blue staining. Deletions of either the N-terminal extension or spacer, or both, did not affect SRSF3 binding, whereas mutations of the docking groove residues abolished the interaction. SRPK1 was included as a positive control. C, in vitro radioactive kinase assays using SRPK2WT (1.5 μm) or SRPK2DM (1.5 μm) and SRSF3 (500 nm) in the presence of 100 μm [32P]ATP. Samples were resolved by SDS-PAGE and visualized by autoradiography. Mutations of the critical docking groove residues reduce the phosphorylation level of SRSF3 from 9.5 ± 0.5 to 4.1 ± 0.2 sites.