Abstract
Phospholipase B–mediated hydrolysis of phosphatidylcholine (PC) results in the formation of free fatty acids and glycerophosphocholine (GPC) in the yeast Saccharomyces cerevisiae. GPC can be reacylated by the glycerophosphocholine acyltransferase Gpc1, which produces lysophosphatidylcholine (LPC), and LPC can be converted to PC by the lysophospholipid acyltransferase Ale1. Here, we further characterized the regulation and function of this distinct PC deacylation/reacylation pathway in yeast. Through in vitro and in vivo experiments, we show that Gpc1 and Ale1 are the major cellular GPC and LPC acyltransferases, respectively. Importantly, we report that Gpc1 activity affects the PC species profile. Loss of Gpc1 decreased the levels of monounsaturated PC species and increased those of diunsaturated PC species, whereas Gpc1 overexpression had the opposite effects. Of note, Gpc1 loss did not significantly affect phosphatidylethanolamine, phosphatidylinositol, and phosphatidylserine profiles. Our results indicate that Gpc1 is involved in postsynthetic PC remodeling that produces more saturated PC species. qRT-PCR analyses revealed that GPC1 mRNA abundance is regulated coordinately with PC biosynthetic pathways. Inositol availability, which regulates several phospholipid biosynthetic genes, down-regulated GPC1 expression at the mRNA and protein levels and, as expected, decreased levels of monounsaturated PC species. Finally, loss of GPC1 decreased stationary phase viability in inositol-free medium. These results indicate that Gpc1 is part of a postsynthetic PC deacylation/reacylation remodeling pathway (PC-DRP) that alters the PC species profile, is regulated in coordination with other major lipid biosynthetic pathways, and affects yeast growth.
Keywords: phosphatidylcholine, phospholipid metabolism, acyltransferase, yeast, membrane turnover, Ale1, glycerophosphocholine, Gpc1, lipid remodeling
Introduction
Cellular membranes must adjust their lipid composition in response to internal and external cues. These adjustments occur through the coordinated control of multiple metabolic activities, including biosynthetic enzymes involved in lipid synthesis, phospholipases involved in lipid turnover, and acyltransferases involved in lipid remodeling. Because lipid composition affects the biophysical properties of the membrane, these alterations are crucial to cellular function (1, 2). Defects in lipid metabolism are associated with multiple disease states and cellular dysfunctions (3–8).
Phosphatidylcholine (PC)2 is the major glycerophospholipid in most eukaryotic membranes. In Saccharomyces cerevisiae, bulk synthesis of PC occurs primarily via the PE methylation pathway and the CDP-choline (Kennedy) pathway (Fig. 1). In the absence of exogenous choline, the CDP-choline pathway does not contribute to net PC synthesis but is important for recycling choline derived from turnover pathways. Characterized turnover pathways include the production of phosphatidic acid and free choline via phospholipase D activity (Spo14/Pld1) (9, 10) and the production of free fatty acids and GPC via phospholipases of the B type encoded by NTE1 and PLB1 (11–13). GPC can subsequently be degraded to free choline and glycerol-3-phosphate by the glycerophosphodiesterase, Gde1 (14, 15). Based on in vitro experiments with yeast extracts, another route for GPC conversion, namely its acylation to LPC, was described (16). Recently, a gene encoding such a GPC acyltransferase activity, GPC1, was identified (17). Subsequent to LPC formation by Gpc1, the lysophospholipid acyltransferase, Ale1, can act to form PC (18). Overall, the degradation of PC to form GPC, followed by its stepwise reacylation to PC, defines a novel PC deacylation/reacylation remodeling pathway (PC-DRP) for PC biosynthesis (12, 17, 19).
Figure 1.
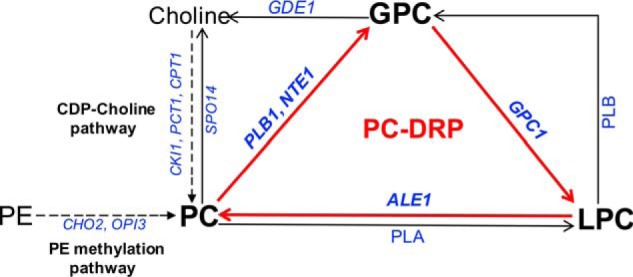
Schematic outline of PC metabolism in yeast. The PC-DRP is indicated by red arrows. The PC-DRP includes deacylation of PC to GPC by PLB1 or NTE1 followed by stepwise reacylation of GPC to LPC by GPC1 and LPC to PC by ALE1. The de novo PC synthesis routes are indicated by dotted arrows. Gene names are in italics. PLA, phospholipase A; PLB, phospholipase B.
PC, like other phospholipids, consists of multiple molecular species based on acyl chain differences at the sn-1 and sn-2 position of the glycerol backbone. The repertoire of fatty acids in yeast is relatively simple as compared with the higher eukaryotes, consisting primarily of C16 and C18 fatty acids with either one or no double bond (20). Thus, the four major PC species observed through MS have the following acyl chain combinations: 32:1PC (monounsaturated) consists of C16:0 and C16:1, 32:2PC (diunsaturated) consists of C16:1 and C16:1, 34:2PC (diunsaturated) consists of C16:1 and C18:1, and 34:1PC (monounsaturated) consists of either C16:0 and C18:1 (this being the predominant acyl chain combination) or C16:1 and C18:0 (21–23).
The steady-state PC molecular species profile is the result of the molecular specificity of the PC biosynthetic routes combined with post synthetic acyl chain exchange (21). The PE methylation pathway produces predominantly diunsaturated PC species, whereas the CDP-choline pathway produces a more mixed profile (21, 24). Furthermore, a number of studies have provided evidence for the postsynthetic remodeling of PC species (20, 25). Indeed, PC remodeling was clearly demonstrated by pulse-chase studies using deuterium-labeled (methyl-D3)-methionine followed by electrospray ionization–MS/MS analysis. Those studies revealed a postsynthetic increase in monounsaturated PC species (32:1 and 34:1) at an expense of diunsaturated PC species (32:2 and 34:2) (24). Prior to this work, an acyltransferase responsible for remodeling PC to more saturated species in S. cerevisiae had not been identified.
Here we undertook a more thorough examination of PC remodeling via PC-DRP and the role of the GPC acyltransferase, Gpc1. Importantly, we report that Gpc1 is a key player in postsynthetic PC remodeling events that result in more saturated PC species. In addition, we find that Gpc1 is regulated with respect to other aspects of lipid metabolism and that loss of Gpc1 impacts stationary phase viability.
Results
GPC can be hydrolyzed by Gde1 or acylated by Gpc1
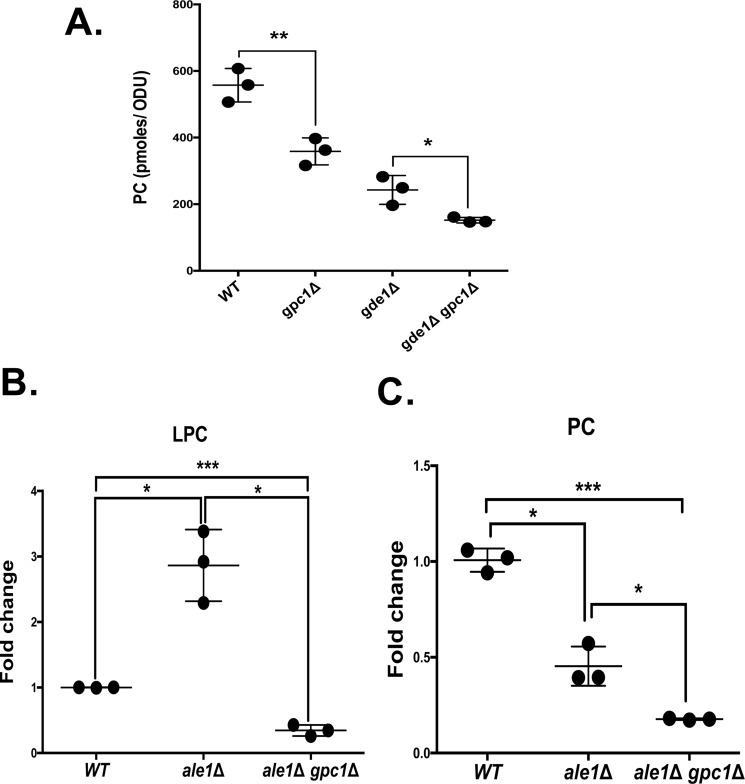
Cellular GPC has two potential metabolic fates. The glycerophosphodiesterase encoded by GDE1 can hydrolyze GPC to produce glycerol-3-phosphate and choline (14, 19). Additionally, GPC can be acylated (likely at the sn-1 position) to LPC by Gpc1, a recently described acyltransferase (17). LPC can be converted to PC by Ale1, consistent with its role as a lysophospholipid acyltransferase (18, 26). To gain further insight into the relative flux of GPC toward hydrolysis versus acylation, in vivo metabolic labeling was employed in strains lacking GPC1 and/or GDE1 (Fig. 2). Cells were grown in the presence of [14C]GPC, which enters through the Git1 transporter (27–29), and label incorporation in the lipid fraction was determined. The [14C]LPC intermediate in the acylation of [14C]GPC to [14C]PC is virtually undetectable under the uniform labeling conditions employed in Fig. 2A; therefore only the label in PC is quantified. As shown in Fig. 2A, a gpc1Δ mutant incorporates roughly 35% less label into PC as compared with WT, consistent with its role as a GPC acyltransferase. A gde1Δ mutant incorporates 50% less label into PC, indicating that a large portion of the label incorporated into PC in a WT strain under low phosphate conditions originates from free [14C]choline released through the action of Gde1. A gde1Δ gpc1Δ double mutant incorporates even less label, confirming the importance of both metabolic routes in determining the ultimate fate of GPC. The label remaining in the gde1Δ gpc1Δ double mutant is likely the result of an uncharacterized acyltransferase and/or glycerophosphodiesterase that can act on GPC.
Figure 2.
The role of Gpc1 in PC metabolism. A, the indicated strains were grown to log phase in the presence of 5 μm [14C]choline-GPC. The cells were harvested, phospholipids were extracted and separated, and PC was quantified as described under “Experimental procedures.” B and C, indicated strains contained plasmid in which GIT1 was constitutively expressed under the control of the ADH1 promoter. The cells were grown in the presence of [14C]choline-GPC for 4 h and harvested, and phospholipids were extracted and analyzed as described under “Experimental procedures.” The data are normalized to the WT containing ADH1-GIT1. Roughly 100-fold more PC was detected as compared with LPC in the WT strain. The data represent the averages of three independent cultures ± S.D. A t test was performed to determine significance as indicated. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005.
Gpc1 and Ale1 are central players in the stepwise acylation of GPC to LPC and then to PC
To demonstrate that Gpc1 produces LPC in vivo and thereby confirm its cellular role, we designed an alternative labeling scheme and employed an ale1Δ mutant to block the second acylation step (Fig. 2B). In the experiment represented in Fig. 2A, cells were grown under low-phosphate conditions to induce the expression of GIT1 (28), a condition that also induces the expression of GDE1 (14, 30). Here, a plasmid harboring GIT1 under the control of the constitutive ADH1 promoter (31) was employed to allow cell growth under normal phosphate level, a condition that represses the expression of GDE1 and therefore reduces the hydrolysis of GPC once it enters the cell (14, 30). In addition, the labeling time in the presence of [14C]GPC was reduced to roughly two generations. Under these conditions, a 3-fold increase in labeled LPC was detected in an ale1Δ mutant as compared with WT, and that increase was counteracted by simultaneous deletion of GPC1 (see the ale1Δ gpc1Δ double mutant). At the same time, labeled PC levels decreased in an ale1Δ mutant and decreased further in an ale1Δ gpc1Δ double mutant. The fact that PC levels were reduced by only 50% in the ale1Δ strain suggests that other enzymes may exist that are capable of converting LPC to PC in the cell. However, an additional possibility is that the increased LPC being produced is hydrolyzed, ultimately releasing free choline that could be incorporated into PC via the Kennedy pathway. These results confirm the formation of the immediate product of Gpc1 activity, LPC, in vivo, and the role of Ale1 in converting LPC into PC.
Gpc1 impacts the PC molecular species profile
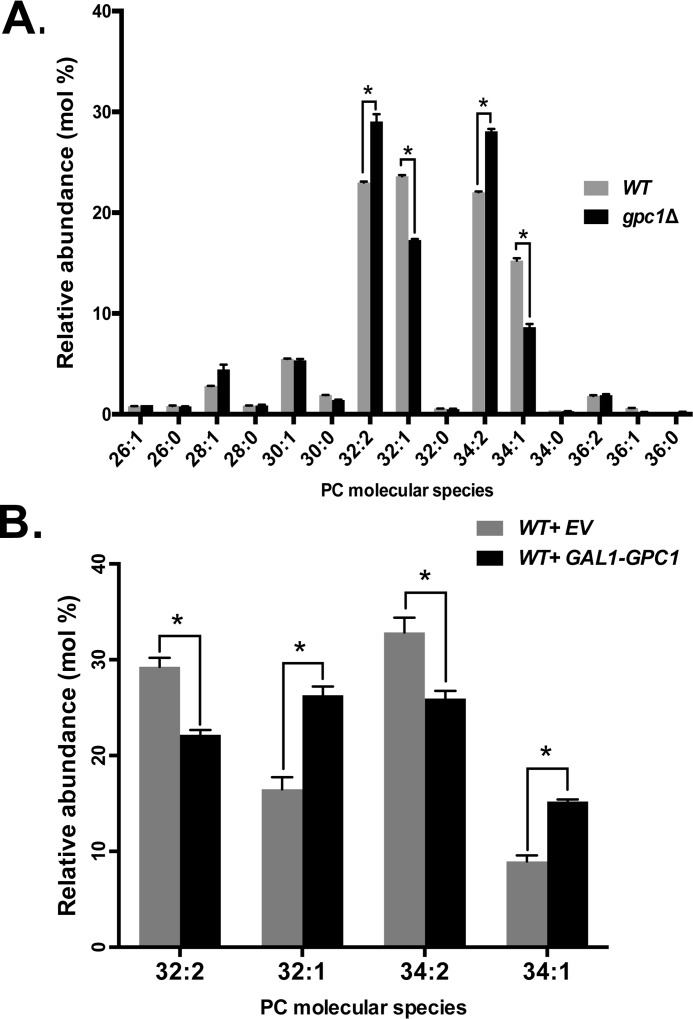
A potential function for the PC-DRP is to modify the acyl chain content. To determine whether Gpc1 activity affects the PC molecular species profile, LC-MS/MS was performed (parent ion scanning for m/z 184). The cells were not supplemented with exogenous GPC for these experiments, so the substrate for Gpc1 activity presumably arose through endogenous phospholipase B activity (19). Fig. 3A includes the range of PC species produced by yeast. Focusing on the major species, the data indicate that the loss of Gpc1 results in a decrease in monounsaturated PC species (32:1, 34:1) and an increase in diunsaturated PC species (32:2, 34:2) as proportions of the total PC pool. Overexpression, as predicted, has the opposite effect (Fig. 3B). When GPC1 was placed under the control of the GAL1 promoter and cells were grown on galactose, there was an increase in monounsaturated PC species (32:1 and 34:1) and a decrease in diunsaturated PC species (32:2, 34:2). In total, these results indicate that Gpc1 impacts the PC species profile and suggest that Gpc1 activity favors the utilization of saturated acyl-CoA species when acylating GPC.
Figure 3.
Gpc1 impacts PC molecular species profile. A and B, the indicated strains were grown to late log phase, the cultures were harvested, and the lipids were extracted. PC species were separated and analyzed using LC-MS/MS, as described under “Experimental procedures.” The data represent averages of three independent cultures ± S.D. A t test was performed to determine significance as indicated. *, p ≤ 0.05.
Loss of Pct1 and Ale1 has minor effects on the PC species profile
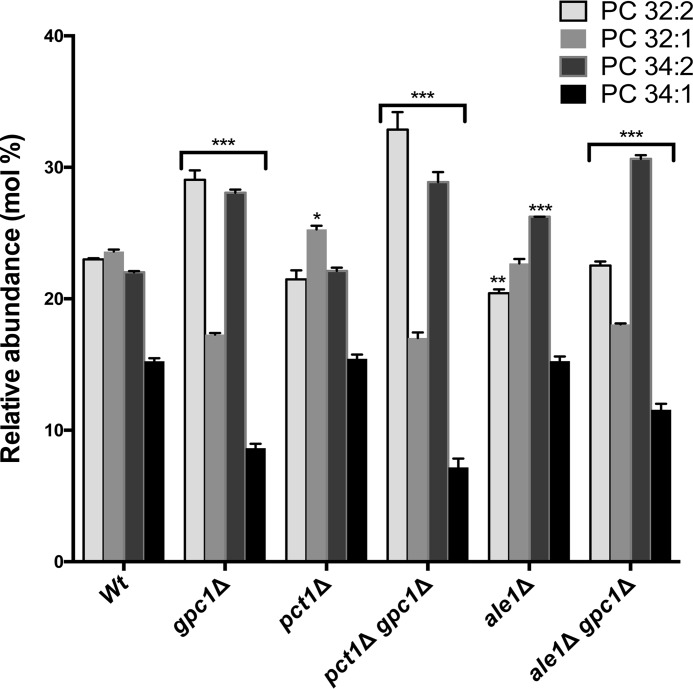
Because Ale1 converts LPC to PC as part of the PC-DRP, we analyzed its impact on the PC molecular species profile. If Ale1 were the only LPC acyltransferase in the cell, and assuming that Ale1 does not affect the species profiles of the lipid precursors of PC (PE and diacylglycerol), the profile of the ale1Δ mutant should resemble that of the gpc1Δ mutant, because the reacylation pathway would stall at LPC, and the LPC produced would likely be rapidly hydrolyzed by phospholipases. In fact, the ale1Δ mutant did not resemble the gpc1Δ strain in terms of PC species, because a slight decrease in PC32:2 and an increase in PC34:2 were its only significant changes from WT. The ale1Δ gpc1Δ mutant, like gpc1Δ, exhibited an increase in diunsaturated species and a decrease in monounsaturated species as compared with WT, although the magnitude of the changes was not identical between the two strains (Fig. 4). These results suggest that there may be more acyltransferases capable of acylating LPC in the cell (32, 33).
Figure 4.
Loss of PCT1 or ALE1 has minor effects on PC species profile. The indicated strains were grown to late log phase, the cultures were harvested, and the lipids were extracted. PC species were separated and analyzed as described under “Experimental procedures.” The data represent averages of three independent cultures ± S.D. A two-way analysis of variance was performed to determine significance for each strain compared with the WT. *, p ≤ 0.05; **, p ≤ 0.004; ***, p ≤ 0.0001.
The steady-state PC molecular species profile in a cell represents a combination of PC biosynthesis, PC degradation, and PC acyl chain remodeling. In the experiments presented in Fig. 3, cells were grown in the absence of free choline, so the PE methylation pathway, not the Kennedy pathway, was primarily responsible for PC biosynthesis. However, flux through the Kennedy pathway can also occur as a result of free choline released through phospholipase-mediated turnover. To examine the extent to which PC turnover resulting in free choline release followed by resynthesis contributes to the observed PC species profile, we utilized a pct1Δ mutant in which flux through the Kennedy pathway is blocked. As shown in Fig. 4, the PC species profile in pct1Δ was largely unchanged from WT, in agreement with Ref. 22, but did result in a small but significant increase in 32:1 PC. That increase may be the result of the observed uptick in GPC1 transcript upon PCT1 deletion (see Fig. 6), as evidenced by the fact that 32:1 again decreases in a pct1Δ gpc1Δ double mutant. Overall, these results indicate that the PC species profiles generated under our experimental conditions are primarily the result of PC synthesis via the PE methylation pathway followed by PC remodeling via PC-DRP.
Figure 6.
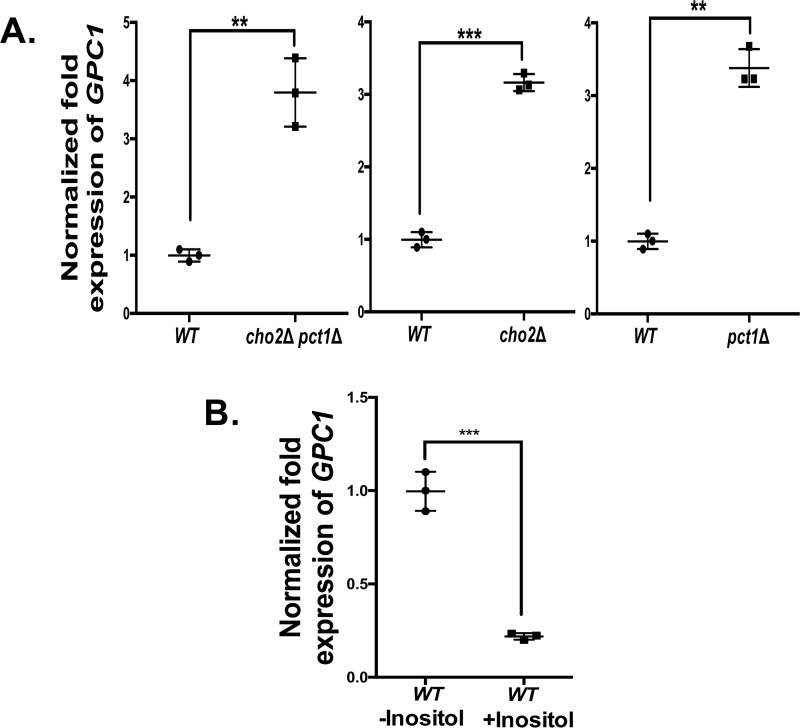
GPC1 transcript is increased by attenuation of PC and by inositol limitation. A and B, the indicated strains were grown to log phase, cells were harvested, and RNA was extracted. GPC1 transcripts were quantified by qRT-PCR. The data were normalized to the amount of the endogenous control mRNA, SNR17, and expressed relative to the WT strain. B, WT grown in the presence of 75 μm inositol (WT +Inositol) compared with WT grown in the absence of inositol (WT −Inositol). The experiments were performed in biological triplicate and assayed in experimental triplicate. A t test was performed to establish significance. **, p ≤ 0.005; ***, p ≤0.0005.
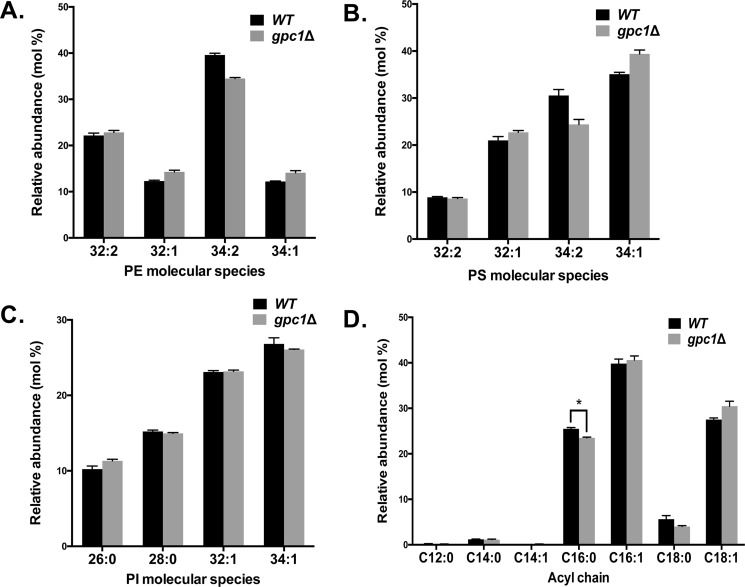
Loss of Gpc1 does not affect PE, PI, and PS species profiles and slightly decreases total 16:0 FA content
We reported previously that Gpc1 had limited ability to utilize acyl-CoAs to acylate glycerophosphoethanolamine in an in vitro system (17). To probe the possibility that Gpc1 acylates glycerophosphoethanolamine to affect PE in vivo, we analyzed the PE molecular species. As shown in Fig. 5A, we did not detect any significant difference in PE molecular species profile when comparing a WT strain to a gpc1Δ strain. Hence, Gpc1 does not play a major role, if at all, in PE remodeling in vivo under the conditions employed here.
Figure 5.
Loss of GPC1 has no significant effects on PE, PS, and PI species profiles and slightly decreases the total C16:0 acyl chain content. The indicated strains were grown to log phase and harvested, and the lipids were extracted. A–C, PE (A), PS (B), and PI (C) species were analyzed by MS as detailed under “Experimental procedures.” D, total acyl chain composition was determined using GC. The data represent averages of four independent cultures ± S.D. A t test was performed to determine significance as indicated. *, p = 0.0008.
The effect of GPC1 deletion on the phosphatidylinositol (PI) and phosphatidylserine (PS) species profiles was also examined. As shown in Fig. 5 (B and C), no significant differences between WT and gpc1Δ were found. In addition, the overall fatty acid composition of all cellular lipids was determined by GC. As shown in Fig. 5D, WT and gpc1Δ fatty acid compositions are largely the same, with the exception of a small but significant decrease in C16:0 in gpc1Δ. This change is consistent with the fact that the gpc1Δ mutant displays a decrease in monounsaturated PC species and may reflect the loss of a small C16:0 sink resulting from the inability of gpc1Δ to add C16:0 to GPC during the PC remodeling process. The results indicate that loss of GPC1 does not induce large-scale changes in fatty acid composition but rather highly specific effects.
GPC1 expression is up-regulated by attenuation of PC biosynthesis and by inositol limitation
To gain further insight into the physiological role of Gpc1, we examined its transcriptional regulation in response to alterations in major lipid biosynthetic pathways. To examine PC biosynthesis, we employed a cho2Δ mutant (attenuated PE methylation pathway), a pct1Δ mutant (blocked Kennedy pathway), and a cho2Δ pct1Δ mutant. The PE methylation pathway for PC biosynthesis is not completely blocked in the cho2Δ pct1Δ double mutant, because of a partially redundant function supplied by the methyltransferase, Opi3 (34). As shown in Fig. 6A, GPC1 message levels, as measured by qRT-PCR, are increased roughly 3-fold in each single mutant and in the double mutant as compared with WT. Thus, inhibition of either pathway for PC biosynthesis causes an increase in GPC1 message, but the effect is not additive.
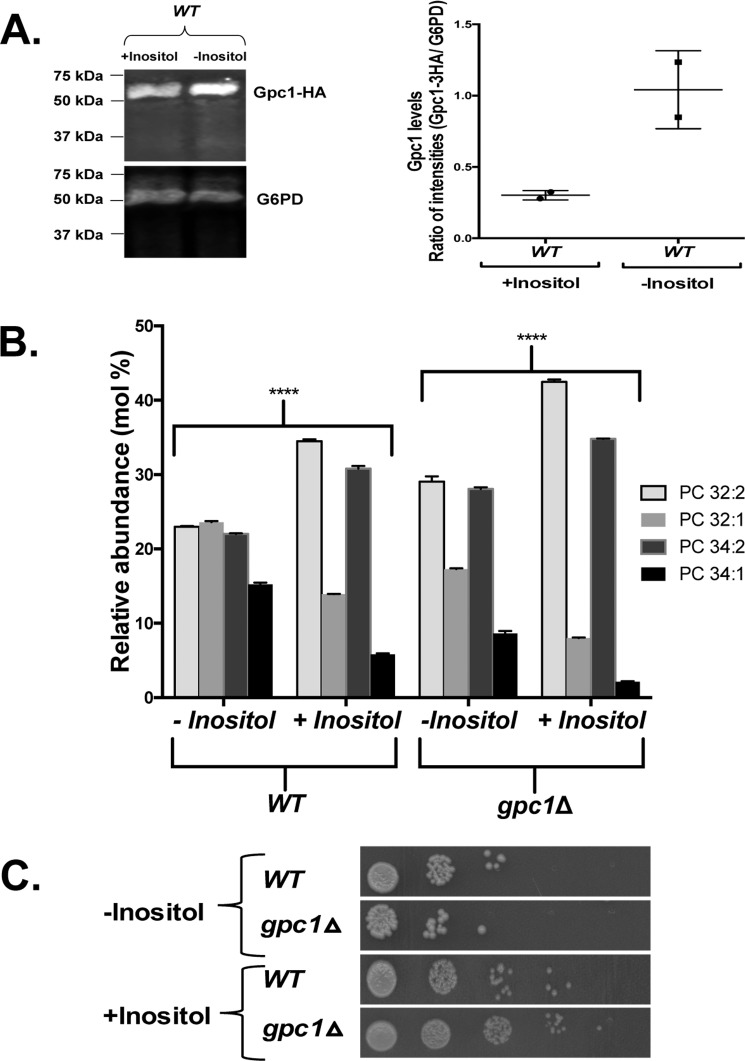
A number of phospholipid biosynthetic genes are regulated by the presence of inositol in the medium via a complex mechanism involving the modulation of phosphatidic acid levels at the ER, which in turn impacts the transit of the Opi1 repressor protein into the nucleus (35, 36). Thus, inositol availability has a general impact on phospholipid composition and flux through the various pathways (10, 35). As shown in Fig. 6B, there is a roughly 5-fold decrease in GPC1 message upon inositol supplementation.
Inositol supplementation represses GPC1 expression and increases diunsaturated PC species
Following the qRT-PCR findings (Fig. 6), the effect of inositol availability was examined more thoroughly, because this perturbation produced the largest transcriptional response and would not confound the interpretation of the PC species profile by impacting PC biosynthesis (as would the cho2Δ and cho2Δ pct1Δ mutants). As seen in Fig. 7A, the decrease in GPC1 transcript upon inositol supplementation was reflected in a decrease in Gpc1 as measured by Western blotting analysis. It should be noted that the PC species profiles presented in Fig. 3 were performed on cells grown in the absence of inositol (I−) to maximize GPC1 expression. When grown in the presence of inositol (I+), there is a clear increase in diunsaturated PC species and a clear decrease in monounsaturated PC species (Fig. 7B) in a WT strain, similar to the effect of GPC1 deletion (Figs. 3A and 7B) in I− medium. A similar pattern of changes in PC molecular species profile upon inositol limitation was reported previously (37). Loss of Gpc1 and inositol supplementation together result in an even greater increase in diunsaturated species, indicating that factors beyond Gpc1 are at play in impacting the PC species profile under these conditions.
Figure 7.
Inositol supplementation affects Gpc1 protein abundance and PC species profile. A, WT strain was grown in medium containing 75 μm inositol (+Inositol) or lacking inositol (−Inositol). Western blotting analysis was performed using anti-HA mouse IRdye 680 and goat anti-mouse IRdye 800. Glucose-6-phosphate dehydrogenase was used as a loading control. Protein bands were quantified using Image StudioTM software. The data represent two biological replicates. B, the indicated strains were grown to late log phase before harvesting. Phospholipids were extracted, and PC species were analyzed using LC-MS/MS. The data represent averages of three independent cultures ± S.D. A t test was performed to establish significance. ****, p ≤ 0.0005. C, strains were cultured in synthetic medium, harvested, and resuspended in sterile water. 10-fold serial dilutions (5 μl) were spotted onto plates containing (+Inositol) or lacking (−Inositol) inositol.
Loss of Gpc1 impacts growth and stationary phase viability in inositol-free medium
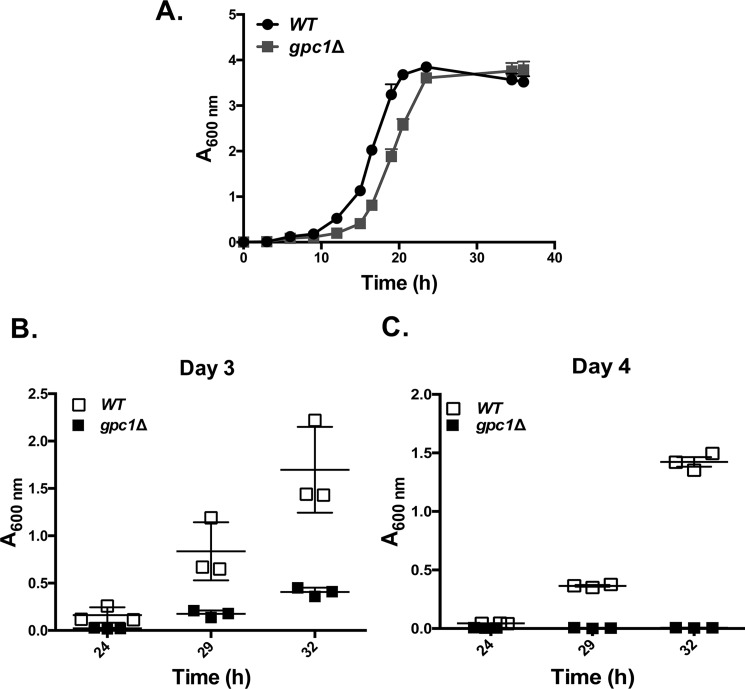
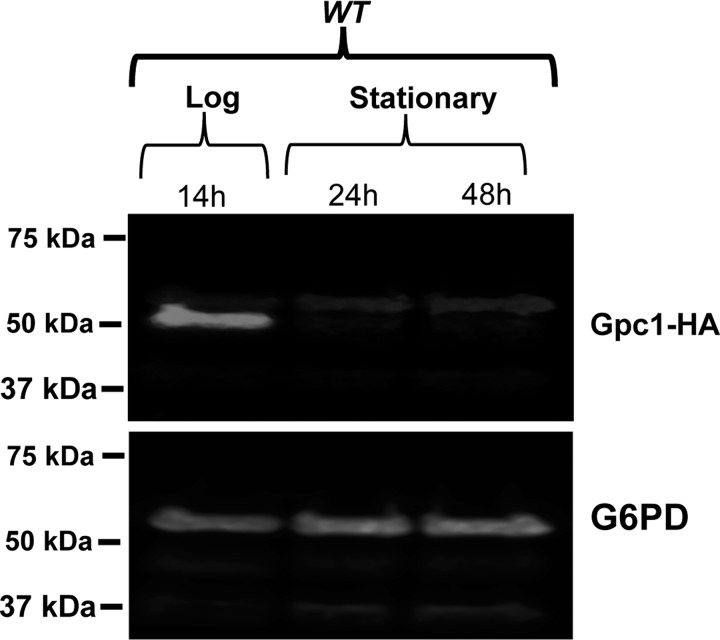
Growth assays were performed to assess the phenotypic consequences of diminished PC remodeling as a function of inositol availability. On plates, gpc1Δ displayed a slight defect in growth in I− conditions, but not I+ conditions (Fig. 7C). This difference was more clearly evident in liquid culture. As shown in Fig. 8A, gpc1Δ mutants displayed an increase in lag time, resulting in slower growth overall, although the rate of doubling during logarithmic phase was comparable with WT. To determine whether decreased ability of cells to regrow from stationary phase was the reason behind the increased lag time, a recultivation experiment was performed. The cells were grown to stationary phase (as in Fig. 8A), but the cultures were incubated longer, for a total of 4 days. On days 3 and 4, the cells were taken from the original cultures and inoculated in fresh YNB I− medium. By day 3, gpc1Δ lagged well behind WT in its ability to reinitiate growth as measured at three time points over 32 h (Fig. 8B; Day 3). In 4-day-old cultures, no growth was detected in the gpc1Δ cultures as measured over 32 h (Fig. 8C; day 4). These results indicate that loss of Gpc1 results in a loss in stationary phase viability in inositol-free medium. Western blotting analysis was performed to examine Gpc1 protein abundance as a function of growth phase. Although Gpc1 was found to be expressed in logarithmic phase (Fig. 9), little or no expression was detected in stationary phase (24 and 48 h of growth). This finding suggests that no further remodeling of PC via this pathway occurs once the cells reach stationary phase.
Figure 8.
Loss of GPC1 impacts growth and stationary phase viability in inositol-free medium. A, indicated strains were grown in synthetic liquid medium lacking inositol, and growth was monitored by measuring optical density at A600 nm over 40 h. The cultures were allowed to continue shaking at 30 °C until day 4 (roughly 96 h). B and C, on days 3 and 4, the cells were taken from the original cultures and inoculated in fresh YNB I− medium to an A600 nm of 0.005. A600 nm was determined at 24, 29, and 32 h. Experiments were performed with three biological replicates (all data points not shown).
Figure 9.
Gpc1 is expressed in logarithmic phase. WT strain was grown in medium lacking inositol. The cultures were harvested at log (14 h), early stationary (24 h), and late stationary (48 h) phases. Western blotting analysis was performed using anti-HA mouse IRdye 680 and goat anti-mouse IRdye 800. Glucose-6-phosphate dehydrogenase was used as a loading control. The experiment was performed in biological triplicate with identical results (no detectable expression in stationary phase).
Discussion
PC metabolism consists of interconnected pathways of synthesis, degradation, recycling of catabolites, and remodeling (Fig. 2A). Deacylation of PC via phospholipases of the B type produces GPC. Our previous studies have shown that GPC is found both intracellularly and extracellularly and that it can be transported into the cell via the Git1 permease (14, 29). A well-characterized pathway for GPC degradation is its hydrolysis to choline and glycerol-3-phosphate by the glycerophosphodiesterase, Gde1 (14, 15). Prior to the identification of GPC1, GPC acyltransferase activity had been detected in Xanthomonas campestris, S. cerevisiae, and plants (16, 38, 39). Our recent identification of the gene encoding Gpc1 provided a tool for examining the relative flux of GPC between degradation versus acylation. When considering the label incorporated into PC from exogenous GPC, roughly 50% occurs via Gde1 activity producing free choline that is subsequently utilized by the Kennedy pathway. Roughly 30% is acylated by Gpc1, and another 20% is presumably metabolized by unknown enzymes. Thus, reacylation is not a trivial pathway for GPC conversion to PC. Through the use of mutants and radiolabeling (Fig. 2B), the primary roles of Gpc1 and Ale1 in the stepwise conversion of GPC to PC as part of the PC-DRP were confirmed.
Although the acylation of GPC to PC clearly occurs in the cell, the synthesis of PC at a level that supports growth requires either a functional PE methylation pathway or a functional Kennedy pathway (25, 40). Hence, we hypothesized that the primary purpose of GPC reacylation may not be bulk synthesis but rather the synthesis of specific PC species as part of a remodeling pathway. Evidence for postsynthetic PC remodeling in yeast has been described by multiple research groups (20, 25, 41–43). Importantly, de Kroon and co-workers (22) have used stable-isotope pulse-chase analysis coupled with MS to prove the postsynthetic formation of more saturated PC species as a function of time and demonstrated the involvement of Plb1. Prior to the identification of Gpc1 activity (16, 17), PC remodeling was believed to involve the removal of a single acyl chain at the sn-2 position, followed by reacylation by a LPC acyltransferase, a cycle first described by Lands (44). Once LPC is formed, it can be acylated to PC by Ale1 (18, 26, 45), with some publications indicating that Ale1 prefers incorporation of an unsaturated acyl-CoA species at the sn-2 position of 1-acyl LPC (18, 45–48). A recent publication has provided evidence that Ale1 may also have limited ability to acylate the sn-1 position of the synthetic LPC analog, 2-alkyl-sn-glycero-3-phosphocholine (43). Importantly, we posit that the previous step in the remodeling pathway, the acylation of GPC, is carried out by the glycerophosphocholine acyltransferase activity of Gpc1, adding another dimension to the process of PC remodeling, namely the potential exchange of both acyl chains. Of course, this finding does not preclude the possibility of PC remodeling occurring via other acyltransferase or transacylase activities (32, 33). PC molecular species profiling of cells lacking GPC1 (Fig. 3A) or overexpressing GPC1 (Fig. 3B) clearly indicates that Gpc1 impacts the PC species profile, producing a higher content of more saturated PC species (32:1 and 34:1) when present. The impact of Gpc1 on phospholipid molecular species is limited to PC, because PI, PE, and PS species profiles are unchanged by loss of Gpc1.
The obvious general question arising from these results is under what circumstances are Gpc1 activity and PC remodeling beneficial? The PC molecular species profile is highly dynamic (49, 50). Given that changes in PC saturation status may well affect a membrane property such as fluidity, we reasoned that Gpc1 must be co-regulated with regards to other lipid metabolic processes to maintain optimal bilayer function. As a first approach, we utilized qRT-PCR to assess changes in transcript levels by attenuating the methylation pathway (cho2Δ) or blocking the Kennedy pathway (pct1Δ) either singly or in combination results in an uptick in GPC1 message levels. Granted, this experiment does not have a straightforward interpretation, because the inhibition of the major PC biosynthetic pathways may cause an increase in GPC reacylation as an end toward preserving PC, not remodeling PC. By similar reasoning, we did not test the effects of choline supplementation in this study (but will be examined in future studies), because that perturbation would increase biosynthesis through the CDP-choline pathway, potentially obscuring the interpretation of the findings. Nonetheless, our results clearly show that the GPC1 message increases upon inhibition of PC biosynthesis.
Inositol availability has far-reaching transcriptional and physiological effects in yeast. Inositol is required for PI biosynthesis. In turn, PI is a precursor for phosphoinositides, polyphosphates, inositol sphingolipids, and glycosylphosphatidylinositol anchors (10, 35, 37). Inositol can be synthesized by the cell and/or imported into the cell from the medium (51). In the absence of exogenous inositol, a number of genes involved in phospholipid metabolism are transcriptionally induced (10, 35). Inositol supplementation results in a 5-fold down-regulation of GPC1 message and a 3-fold decrease in protein level (Figs. 6B and 7A). Down-regulation of GPC1 transcript and protein levels by inositol supplementation resulted in changes in the PC species profile, providing an explanation for a similar result reported by Henry and colleagues (37) prior to the identification of Gpc1.
Most genes responsive to inositol limitation contain one or more UASINO elements (consensus sequence 5′-CATGTGAAAT-3′) in their promoters and are regulated by Ino2, Ino4, and Opi1 via the Henry Regulatory Circuit (35, 52, 53). The transcription factors, Ino2 and Ino4, typically bind to UASINO as a heterodimer to activate transcription when inositol is limiting, and the Opi1 repressor binds to Ino2 to block transcriptional activation when inositol is available. The Gpc1 promoter contains a sequence (CTTGTGAATA) that is 3 bp off from the consensus Ino2–Ino4 binding sequence (54) at −193 to −202. In addition, it contains a site (CATTTG) for Ino4-dependent/Ino2-independent binding (55) at −182 to −187 bp. Further evidence for control by the Henry Regulatory Circuit is provided by microarray studies performed prior to the characterization of Gpc1 in which Opi1 (56) and Ino4 (57) were shown to affect transcription of the GPC1 ORF, YGR194w. Further studies would be required to unequivocally prove binding of Ino2 and/or Ino4 to the GPC1 promoter.
In media lacking inositol, where GPC1 is up-regulated, gpc1Δ mutants display decreased stationary phase viability (Fig. 8). However, we did not detect Gpc1 expression in the stationary phase, suggesting that the alteration in PC species that occurs via PC-DRP during log phase may prepare the membrane for events that occur in later stages of growth. Gpc1 is predicted to be an integral membrane protein with eight transmembrane domains (17). A definitive localization study has not been performed for Gpc1 but will be the focus of future studies and may shed light on its role in cell viability. Of note, the changes in lipid metabolism resulting from I− medium are accompanied by the induction of multiple stress-response pathways, including the unfolded protein response, the protein kinase C–mitogen-activated protein kinase pathway, and others (35). Thus, the loss of cell viability observed under I− conditions upon loss of GPC1 is likely the result of a complex series of events. In summary, our data indicate that Gpc1 is part of PC-DRP, a remodeling pathway that provides the opportunity for postsynthetic alteration of both acyl chains species and, thereby, to impact the biophysical properties of the membrane.
Experimental procedures
Strains, plasmid, media, and growth conditions
The S. cerevisiae strains used in this study are presented in Table 1. Strains were maintained aerobically at 30 °C with shaking or on a roller drum. Growth was monitored by measuring optical densities at 600 nm (A600 nm) using a BioMate3 Thermo Spectronic spectrophotometer. The media used in this study were yeast peptone dextrose or yeast nitrogen base (YNB) with 2% glucose and amino acid composition as described in Ref. 58. The YNB medium was made according to DIFCO manual but lacking inositol. For studies requiring inositol, 75 μm of inositol was provided. Transformations of autonomously replicating plasmids were performed using the lazy bones plasmid transformation protocol as described (59). For integrating DNA into the genome, the high-efficiency transformation procedure was used (60). The empty vector, pRS426, and vector overexpressing GIT1 gene ADH-GIT1 were from Ref. 31.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference | |
|---|---|---|---|
| JPV 399 | WT, BY4742 | BY4742; MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| JPV 788 | gpc1Δ | BY4742; gpc1::KanMX | This study |
| JPV 125 | gde1Δ | BY4742; gde1::KanMX | This study |
| JPV 834 | gde1Δ gpc1Δ | JPV 125; gpc1::URA3 | This study |
| JPV 272 | cho2Δ pct1Δ | BY4742; cho2::KanMX pct1::LEU2 | This study |
| JPV 839 | cho2Δ pct1Δ gpc1Δ | JPV 272; gpc1::URA3 | This study |
| JPV 841 | pct1Δ gpc1Δ | JPV 788; pct1::LEU2 | This study |
| JPV 269 | cho2Δ | BY4742; cho2::KanMX | Research Genetics |
| JPV 270 | pct1Δ | BY4742; pct1::LEU2 | Research Genetics |
| JPV 832 | ale1Δ | Mat a: his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ale1::KANMX | Ref. 17 |
| JPV 833 | ale1Δ gpc1Δ | JPV 832; gpc1::KANMX | Ref. 17 |
| JPV 846 | WT + GPC1-3xHA | BY4742; GPC1–3xHA | This study |
Construction of deletion strains
The WT (BY4742) S. cerevisiae strain was purchased from Open Biosystems (Thermo Scientific, Huntsville, AL). The deletion strains used in this study were made using PCR-based homologous recombination technique as described in Refs. 61 and 62). Nutritional markers, LEU2 and URA3, were amplified from pRS415 and pRS416, respectively, using primers listed in Table 2.
Table 2.
Nucleotide primer sequences used for gene deletions
The bold sequences are homologous to the template plasmid. The deleted gene was replaced with gene name in parentheses.
| Gene name | Primer | Sequence (5′ → 3′) |
|---|---|---|
| GPC1 (URA3) | Forward | ATGTACAAGTTGGACAATAACGACATTGACGATGAAACGAATAACTCTGTTTCACTGACGAGCGAACAAAAGCTGG |
| GPC1 (URA3) | Reverse | CTTAATCACTCTTTGAGGATACACTTGAAGAGTCGTCAAAATCTTCGTCACGGTTGACATCTGTAGGGCGAATTGGG |
| GPC1 (LEU2) | Forward | ATGTACAAGTTGGACAATAACGACATTGACGATGAAACGAATAACTCTGTTTCACTGACGCACATACCTAATATTATTGC |
| GPC1 (LEU2) | Reverse | CTTAATCACTCTTTGAGGATACACTTGAAGAGTCGTCAAAATCTTCGTCACGGTTGACATGAATCTTTTTAAGCAAGGAT |
| GPC1-1-3XHA | Forward | TTGAATGTCAACCGTGACGAAGATTTTGACGACTCTTCAAGTGTATCCTCAAAGAGTGATAGGGAACAAAAGCTGGAG |
| GPC1-1-3XHA | Reverse | GTAAAGCGCCTGTAAATAAAAGCTCTCAAAGTTAACAGATAAATGAAGTGAACTATCTTACTGTAGGGCGAATTGGG |
Construction of chromosomal C-terminal 3×HA tag for GPC1
The 3×HA-URA3 cassette was amplified from plasmid pPMY-3×HA (63) using the primer set shown in Table 2. The PCR product was transformed into WT strain, and transformants were selected on YNB plates lacking uracil. Control PCR was performed on genomic DNA extracted from transformants to verify the integration of the entire cassette into the genome. To counterselect against the URA3 gene, the verified colonies were then reselected on a YNB plate containing 1 mg/ml of 5-fluoroorotic acid. Genomic DNA was extracted from colonies that grew on 5-fluoroorotic acid plates and were used as template to check for the insertion of 3×HA at the C-terminal end. Colonies producing the correct amplicon are JPV846 (WT+GPC1–3×HA) used in the Western blotting analyses.
In vivo labeling and lipid isolation
For the long-term labeling experiment represented in Fig. 2A, strains were grown in YNB inositol-free medium containing 5 μm of [14C]GPC (≅200,000 cpm/ml) (American Radiolabeled Chemicals 3880) and a low concentration of KH2PO4 (200 μm) to induce expression of the Git1 transporter for GPC uptake. Cultures were allowed to grow to logarithmic phase and harvested, and the cell pellets were treated with 5% TCA for 20 min on ice. Following centrifugation, the supernatant was discarded, and cell pellets were incubated at 60°C for 60 min with 1 ml of ESOAK (95% ethanol, diethylether, H2O, pyridine, NH4OH (28–30%); 15:5:15:1:0.036 v/v/v/v/v). The tubes were centrifuged to pellet the debris, and 1 ml of lipid-containing supernatant was transferred to fresh tubes containing 2.5 ml of chloroform/methanol (2:1) and 0.25 ml of 0.1 m HCl. Following vortexing and low-speed centrifugation, the bottom layers containing glycerophospholipids were dried under N2. Lipids were then suspended in chloroform/methanol (2:1) and spotted onto silica gel TLC plates (Whatman, Millipore Sigma 105626), and plates were developed in chloroform:methanol:acetic acid:water; (85:15:10:3.5 v/v/v/v) (39). The TLC plates were imaged using a Typhoon 8200 phosphorimager. For Fig. 2B, ImageQuant software was used to quantitate the PC and LPC spots. For Fig. 2A, no LPC was detected, allowing the radioactivity (cpm) in the lipid fraction to be converted to pmol PC/ODU based on the specific activity of the exogenous label.
For the experiments aimed at detecting LPC (Fig. 2B), strains containing either empty vector or ADH-GIT1 (31) were grown in YNB inositol-free medium containing 5 μm of [14C]GPC (≅200,000 cpm/ml) and a standard level of KH2PO4. The cultures were inoculated at an A600 nm of ∼0.2 from startup cultures. Cultures were allowed to grow for 4 h before harvesting ∼3.2 ODUs of cells for each strain. Lipid extraction, separation, and quantitation were as described in previous paragraph. LPC and PC standards (Avanti Polar lipids) were used to verify lipid migration on the TLC plates. Radiolabeled GPC was purchased from American Radiolabeled Chemicals, and unlabeled GPC was from Sigma Scientific.
Analysis of PC and PE molecular species profiles by MS
The indicated strains were grown to logarithmic phase, and 30 ODUs of each were harvested. The cell pellets were suspended in 2 ml of 5% TCA, followed by a 15-min incubation on ice. After centrifugation, the cell pellets were washed with 20 ml of Nano-pure H2O. Phospholipids were extracted from the pellet as described above and were finally resuspended in 500 μl of CHCl3:MeOH (1:1, v/v). A 5-μl lipid sample was injected into an Agilent 1200 HPLC system coupled to an Agilent 6460 triple quadrupole LC/MS system. Lipid separations were performed on a Varian PLRP-S (150 × 2.1 mm, 5 μm) column at a flow rate of 0.35 ml/min, and the column was maintained at 50 °C. Samples were eluted with a linear gradient using of 75% methanol in water (A) and 100% methanol (B). Both solvents contained 10 mm ammonium acetate and 0.1% formic acid. Electrospray ionization–MS was performed with scanning mode set to multiple reaction monitoring set at m/z 184.1 product ion to detect PC molecules. PE species were detected by neutral loss scanning at 141 atomic mass units in positive ion mode. The MS parameters were optimized at: collision energy, 25 eV for PC and 15 eV for PE; capillary voltage, 3500 V; fragmentor voltage, 100 V; dwell time, 200 ms; drying gas temperature, 320 °C; sheath gas temperature, 350 °C; sheath gas flow, 11 liters/min and drying gas flow, 10 liters/min. The data were acquired and analyzed using Agilent Mass Hunter Work Station software. Reported values for various PC and PE species were corrected for isotope effect (64).
Analysis of PI and PS molecular species profiles by MS
Total lipid extracts were dissolved in CHCl3:MeOH 1:1 and injected in a Ultra-High-Performance Liquid Chromatography (UHPLC) system equipped with a Kinetex HILIC column (50 × 4.60 mm, 2.6 μm; Phenomenex, Utrecht, The Netherlands) with a SecurityGuard ULTRA HILIC precolumn. Lipids were eluted using a binary gradient of acetonitrile:acetone 9:1 (v/v) (with 0.1% formic acid; eluent A) and 50 mm ammonium formate in acetonitrile:H2O 7:3 (v/v) (with 0.1% formic acid; eluent B). The linear gradient was from 100% A to 50% A in 1 min, followed by isocratic elution at the latter composition for an additional 2 min before a 1-min cleaning of the column with 100% B. The flow rate throughout the analysis was 1 ml/min. MS analysis was performed on an LTQ-XL mass spectrometer (Thermo Scientific) using electrospray ionization and detection in the negative mode (source voltage −4.2 kV). Full-scan mass spectra were recorded over a range of 450–950 m/z. The data were converted to mzML format and analyzed using XCMS version 1.52.0 (65) running under R version 3.4.3, including isotope correction.
Analysis of acyl chain composition
Total cellular acyl chain composition was analyzed as described previously (66). Briefly, total lipid extracts corresponding to 100 nmol phospholipid phosphorus were dried, and transesterified in methanolic H2SO4 (40:1 v/v) at 70 °C. Fatty acid methyl esters (FAMEs) were extracted with hexane and separated on a Trace GC (Interscience, Breda, NL) equipped with a biscyanopropyl-polysiloxane column (Restek, Bellefonte, PA). FAMEs were identified, and signal intensity was calibrated using a commercially available standard (Nu-Chek, Elysian, MN). Acyl chain compositions are presented as mol % of total FAMEs recovered.
RNA extraction and qRT-PCR
RNA was extracted from logarithmic phase cells using the hot phenolic RNA extraction protocol (67). The extracted RNA sample was quantified using Thermo Scientific NanoDrop UV-visible spectrophotometry. DNase treatment was performed on 7 μg of RNA using the TURBO DNA-freeTM kit (Applied Biosystems) as per the manufacturer's protocol. The VersoTM SYBR Green one-step QRT-PCR ROX kit (Thermo Scientific) was used to determine the message levels on an Applied Biosystems StepOne-PlusTM real-time PCR thermocycler. The primer nucleotide sequence sets used in these studies are shown in Table 3. The optimal primer concentration (70–100 nm) and template concentration (100 ng/25 μl reaction) were determined by performing a standard curve on each primer set. Reverse transcription was performed at 50 °C for 15 min followed by 95 °C for 15 min for RT inactivation and polymerase activation. The thermal profile for amplification were 40 cycles at 95 °C for 15 s, 54.5 °C for 30 s, and 72 °C for 40 s (62). Primer set specificity was determined by melting curve analysis. A no-template control and a no-RT control were used to validate the absence of contaminants in RNA samples. Experiments were performed using three independent biological replicates, and each replicate was analyzed in experimental triplicates. The data were normalized to the endogenous control gene, SNR17, and analyzed by the ΔΔCt method (68, 69).
Table 3.
Nucleotide primer sequences for qRT-PCR
| Gene name | Primer | Sequence (5′ → 3′) |
|---|---|---|
| SNR17 | Forward | TTG ACT CTT CAA AAG AGC CAC TGA |
| SNR17 | Reverse | CGG TTT CTC ACT CTG GGG TAC |
| GPC1 | Forward | AAA TTA GCT GCG GCC CTA TT |
| GPC1 | Reverse | TAT CAT TCA CGG AGG CAT CA |
Protein extraction and Western blotting analysis
For Fig. 7A, the cells were grown in YNB medium containing or lacking 75 μm of inositol. Cultures corresponding to 25 ODUs of cells were harvested at log phase. The cell pellet was resuspended in ice-cold extraction buffer (120 mm NaCl, 50 mm Tris-HCl, pH 7.5, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1% Nonidet P-40, 0.1% SDS, and 1% Triton X-100) containing 1× Yeast/Fungal Protease ArrestTM mixture (G-Biosciences catalog no. 786-333). Lysis was performed using glass bead for eight cycles of the following: 30 s of vortexing and 30 s of incubation on ice. The cell debris was pelleted by centrifugation at 2000 × g for 5 min. For Fig. 9, the cells were grown in YNB medium lacking inositol. Equivalent ODUs of cells were harvested at log and stationary (24 h) and late stationary phases (48 h). The cell were lysed for 10 min on ice with 50 μl of extraction buffer (0.2 m NaOH, 0.2% mercaptoethanol) containing 1× Yeast/Fungal Protease ArrestTM mixture. Proteins were precipitated with 50 μl of 50% TCA and pelleted at 12,000 × g for 5 min. The pellets were suspended in 120 μl of dissolving buffer (4% SDS, 0.1 m Tris-HCl, pH 6.8, 4 mm EDTA, 20% glycerol, 2% 2-mercaptoethanol, 0.02% bromphenol blue) (70). The samples were heated at 37 °C for 15 min. Protein in the supernatant was quantified using the Bradford assay kit (Thermo Fisher). An equivalent amount of protein was added to each lane. Western blotting analysis was performed as described in Ref. 17. Mouse monoclonal anti-HA (Sigma–Aldrich, catalog no. H3663) was used as primary antibody, and goat anti-mouse antibody (LI-COR, catalog no. 925-32210) was used for detection. The G6PD was used as the loading control (17).
Recultivation from a stationary phase culture
Indicated strains were grown in YNB medium lacking inositol. Cultures were restarted at an A600 nm ∼0.005, and growth curves were determined by measuring optical density at 600 nm (A600 nm) as described previously for 4 days (all data points not shown). The ability to grow following recultivate was examined by restarting the cultures at A600 nm ∼0.005 from day 3 and day 4 of the cultures used in the growth curve. The stationary phase was confirmed by microscopy, where most cells were in the unbudded G0 phase. Growth of the recultivated cultures was monitored at three different time points.
Statistical analysis
Paired t test analysis or two-way analysis of variance was performed to establish significances using GraphPad Prism 6.
Author contributions
S. A., I. L., and J. P.-V. conceptualization; S. A. and J. P.-V. resources; S. A., A. I. P. M. d. K., and J. P.-V. supervision; S. A. and J. P.-V. funding acquisition; S. A., B. J., M. F. R., I. L., and J. P.-V. investigation; S. A. and J. P.-V. writing-original draft; S. A. and J. P.-V. project administration; S. A., R. K., B. J., M. F. R., J. F. H. M. B., I. L., A. I. P. M. d. K., and J. P.-V. writing-review and editing; R. K. and A. I. P. M. d. K. formal analysis; R. K. validation; R. K. and J. F. H. M. B. methodology.
Acknowledgments
The purchase of mass spectrometers was supported by National Science Foundation Grant MRIDBI-0821401 to Duquesne University. We thank Dr. Amrah Weijn for help with the MS sample preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant R15 GM 104876 (to J. P.-V.), PlantLink and the Strategic Research Area Project “Trees and Crops for the Future” (to I. L.), and The Netherlands Organization for Scientific Research (to M. F. R. and A. I. P. M. d. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PC
- phosphatidylcholine
- GPC
- glycerophosphocholine
- LPC
- lysophosphatidylcholine
- PC-DRP
- phosphatidylcholine deacylation/reacylation remodeling pathway
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- YNB
- yeast nitrogen base
- HA
- hemagglutinin
- FAME
- fatty acid methyl ester
- qRT-PCR
- real-time quantitative RT-PCR
- ODU
- optical density unit
- LC-MS/MS
- liquid chromatography with tandem mass spectrometry.
References
- 1. Ernst R., Ejsing C. S., and Antonny B. (2016) Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 428, 4776–4791 10.1016/j.jmb.2016.08.013 [DOI] [PubMed] [Google Scholar]
- 2. Renne M. F., and de Kroon A. (2018) The role of phospholipid molecular species in determining the physical properties of yeast membranes. FEBS Lett. 592, 1330–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laplante M., and Sabatini D. M. (2009) An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 19, R1046–R1052 10.1016/j.cub.2009.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee J. C., Simonyi A., Sun A. Y., and Sun G. Y. (2011) Phospholipases A2 and neural membrane dynamics: implications for Alzheimer's disease. J. Neurochem. 116, 813–819 10.1111/j.1471-4159.2010.07033.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hishikawa D., Hashidate T., Shimizu T., and Shindou H. (2014) Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 55, 799–807 10.1194/jlr.R046094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marchan R., Lesjak M. S., Stewart J. D., Winter R., Seeliger J., and Hengstler J. G. (2012) Choline-releasing glycerophosphodiesterase EDI3 links the tumor metabolome to signaling network activities. Cell Cycle 11, 4499–4506 10.4161/cc.22544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cornell R. B., and Ridgway N. D. (2015) CTP:phosphocholine cytidylyltransferase: function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog. Lipid Res. 59, 147–171 10.1016/j.plipres.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 8. Fagone P., and Jackowski S. (2013) Phosphatidylcholine and the CDP-choline cycle. Biochim. Biophys. Acta, 1831, 523–532 10.1016/j.bbalip.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sreenivas A., Patton-Vogt J. L., Bruno V, Griac P, and Henry S. A. (1998) A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J. Biol. Chem. 273, 16635–16638 10.1074/jbc.273.27.16635 [DOI] [PubMed] [Google Scholar]
- 10. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee K. S., Patton J. L., Fido M., Hines L. K., Kohlwein S. D., Paltauf F., Henry S. A., and Levin D. E. (1994) The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269, 19725–19730 [PubMed] [Google Scholar]
- 12. Patton-Vogt J. (2007) Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim. Biophys. Acta 1771, 337–342 10.1016/j.bbalip.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 13. Fernández-Murray J. P., Gaspard G. J., Jesch S. A., and McMaster C. R. (2009) NTE1-encoded phosphatidylcholine phospholipase B regulates transcription of phospholipid biosynthetic genes. J. Biol. Chem. 284, 36034–36046 10.1074/jbc.M109.063958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher E., Almaguer C., Holic R., Griac P., and Patton-Vogt J. (2005) Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280, 36110–36117 10.1074/jbc.M507051200 [DOI] [PubMed] [Google Scholar]
- 15. Fernández-Murray J. P., and McMaster C. R. (2005) Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J. Biol. Chem. 280, 38290–38296 10.1074/jbc.M507700200 [DOI] [PubMed] [Google Scholar]
- 16. Stålberg K., Neal A. C., Ronne H., and Ståhl U. (2008) Identification of a novel GPCAT activity and a new pathway for phosphatidylcholine biosynthesis in S. cerevisiae. J. Lipid Res. 49, 1794–1806 10.1194/jlr.M800129-JLR200 [DOI] [PubMed] [Google Scholar]
- 17. Glab B., Beganovic M., Anaokar S., Hao M. S., Rasmusson A. G., Patton-Vogt J., Banaś A., Stymne S., and Lager I. (2016) Cloning of glycerophosphocholine acyltransferase (GPCAT) from fungi and plants: a novel enzyme in phosphatidylcholine synthesis. J. Biol. Chem. 291, 25066–25076 10.1074/jbc.M116.743062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riekhof W. R., Wu J., Gijón M. A., Zarini S., Murphy R. C., and Voelker D. R. (2007) Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 282, 36853–36861 10.1074/jbc.M706718200 [DOI] [PubMed] [Google Scholar]
- 19. Dowd S. R., Bier M. E., and Patton-Vogt J. L. (2001) Turnover of phosphatidylcholine in Saccharomyces cerevisiae: the role of the CDP-choline pathway. J. Biol. Chem. 276, 3756–3763 10.1074/jbc.M003694200 [DOI] [PubMed] [Google Scholar]
- 20. Wagner S., and Paltauf F. (1994) Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae: fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast 10, 1429–1437 10.1002/yea.320101106 [DOI] [PubMed] [Google Scholar]
- 21. Renne M. F., Bao X., De Smet C. H., and de Kroon A. I. (2015) Lipid acyl chain remodeling in yeast. Lipid Insights 8, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Smet C. H., Cox R., Brouwers J. F., and de Kroon A. I. (2013) Yeast cells accumulate excess endogenous palmitate in phosphatidylcholine by acyl chain remodeling involving the phospholipase B Plb1p. Biochim. Biophys. Acta 1831, 1167–1176 10.1016/j.bbalip.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 23. Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., and Shevchenko A. (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141 10.1073/pnas.0811700106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boumann H. A., Damen M. J., Versluis C., Heck A. J., de Kruijff B., and de Kroon A. I. (2003) The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species: evidence for lipid remodeling. Biochemistry 42, 3054–3059 10.1021/bi026801r [DOI] [PubMed] [Google Scholar]
- 25. Tanaka K., Fukuda R., Ono Y., Eguchi H., Nagasawa S., Nakatani Y., Watanabe H., Nakanishi H., Taguchi R., and Ohta A. (2008) Incorporation and remodeling of extracellular phosphatidylcholine with short acyl residues in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1781, 391–399 10.1016/j.bbalip.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 26. Riekhof W. R., Wu J., Jones J. L., and Voelker D. R. (2007) Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 28344–28352 10.1074/jbc.M705256200 [DOI] [PubMed] [Google Scholar]
- 27. Almaguer C., Fisher E., and Patton-Vogt J. (2006) Posttranscriptional regulation of Git1p, the glycerophosphoinositol/glycerophosphocholine transporter of Saccharomyces cerevisiae. Curr. Genet. 50, 367–375 10.1007/s00294-006-0096-8 [DOI] [PubMed] [Google Scholar]
- 28. Almaguer C., Mantella D., Perez E., and Patton-Vogt J. (2003) Inositol and phosphate regulate GIT1 transcription and glycerophosphoinositol incorporation in Saccharomyces cerevisiae. Eukaryot. Cell 2, 729–736 10.1128/EC.2.4.729-736.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patton-Vogt J. L., and Henry S. A. (1998) GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149, 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng B., Berrie C. P., Corda D., and Farquhar M. G. (2003) GDE1/MIR16 is a glycerophosphoinositol phosphodiesterase regulated by stimulation of G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 100, 1745–1750 10.1073/pnas.0337605100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wykoff D. D., and O'Shea E. K. (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jasieniecka-Gazarkiewicz K., Demski K., Lager I., Stymne S., and Banaś A. (2016) Possible role of different yeast and plant lysophospholipid:acyl-CoA acyltransferases (LPLATs) in acyl remodelling of phospholipids. Lipids 51, 15–23 10.1007/s11745-015-4102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamashita A., Hayashi Y., Nemoto-Sasaki Y., Ito M., Oka S., Tanikawa T., Waku K., and Sugiura T. (2014) Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 53, 18–81 10.1016/j.plipres.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 34. Kodaki T., and Yamashita S. (1987) Yeast phosphatidylethanolamine methylation pathway: cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262, 15428–15435 [PubMed] [Google Scholar]
- 35. Henry S. A., Gaspar M. L., and Jesch S. A. (2014) The response to inositol: Regulation of glycerolipid metabolism and stress response signaling in yeast. Chem. Phys. Lipids. 180, 23–43 10.1016/j.chemphyslip.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hofbauer H. F., Schopf F. H., Schleifer H., Knittelfelder O. L., Pieber B., Rechberger G. N., Wolinski H., Gaspar M. L., Kappe C. O., Stadlmann J., Mechtler K., Zenz A., Lohner K., Tehlivets O., Henry S. A., and Kohlwein S. D. (2014) Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids. Dev. Cell 29, 729–739 10.1016/j.devcel.2014.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaspar M. L., Aregullin M. A., Jesch S. A., and Henry S. A. (2006) Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281, 22773–22785 10.1074/jbc.M603548200 [DOI] [PubMed] [Google Scholar]
- 38. Moser R., Aktas M., and Narberhaus F. (2014) Phosphatidylcholine biosynthesis in Xanthomonas campestris via a yeast-like acylation pathway. Mol. Microbiol. 91, 736–750 10.1111/mmi.12492 [DOI] [PubMed] [Google Scholar]
- 39. Lager I., Glab B., Eriksson L., Chen G., Banaś A., and Stymne S. (2015) Novel reactions in acyl editing of phosphatidylcholine by lysophosphatidylcholine transacylase (LPCT) and acyl-CoA:glycerophosphocholine acyltransferase (GPCAT) activities in microsomal preparations of plant tissues. Planta 241, 347–358 10.1007/s00425-014-2184-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klug L., and Daum G. (2014) Yeast lipid metabolism at a glance. FEMS Yeast Res. 14, 369–388 10.1111/1567-1364.12141 [DOI] [PubMed] [Google Scholar]
- 41. Lands W. E. (1960) Metabolism of glycerolipids: 2. the enzymatic acylation of lysolecithin. J. Biol. Chem. 235, 2233–2237 [PubMed] [Google Scholar]
- 42. Kishino H., Eguchi H., Takagi K., Horiuchi H., Fukuda R., and Ohta A. (2014) Acyl-chain remodeling of dioctanoyl-phosphatidylcholine in Saccharomyces cerevisiae mutant defective in de novo and salvage phosphatidylcholine synthesis. Biochem. Biophys. Res. Commun. 445, 289–293 10.1016/j.bbrc.2014.01.136 [DOI] [PubMed] [Google Scholar]
- 43. Morisada S., Ono Y., Kodaira T., Kishino H., Ninomiya R., Mori N., Watanabe H., Ohta A., Horiuchi H., and Fukuda R. (2018) The membrane-bound O-acyltransferase Ale1 transfers an acyl moiety to newly synthesized 2-alkyl-sn-glycero-3-phosphocholine in yeast. FEBS Lett. 592, 1829–1836 10.1002/1873-3468.13103 [DOI] [PubMed] [Google Scholar]
- 44. Lands W. E. (1965) Lipid metabolism. Annu. Rev. Biochem. 34, 313–346 10.1146/annurev.bi.34.070165.001525 [DOI] [PubMed] [Google Scholar]
- 45. Jain S., Stanford N., Bhagwat N., Seiler B., Costanzo M., Boone C., and Oelkers P. (2007) Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 30562–30569 10.1074/jbc.M706326200 [DOI] [PubMed] [Google Scholar]
- 46. Benghezal M., Roubaty C., Veepuri V., Knudsen J., and Conzelmann A. (2007) SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J. Biol. Chem. 282, 30845–30855 10.1074/jbc.M702719200 [DOI] [PubMed] [Google Scholar]
- 47. Tamaki H., Shimada A., Ito Y., Ohya M., Takase J., Miyashita M., Miyagawa H., Nozaki H., Nakayama R., and Kumagai H. (2007) LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 282, 34288–34298 10.1074/jbc.M704509200 [DOI] [PubMed] [Google Scholar]
- 48. Chen Q., Kazachkov M., Zheng Z., and Zou J. (2007) The yeast acylglycerol acyltransferase LCA1 is a key component of Lands cycle for phosphatidylcholine turnover. FEBS Lett. 581, 5511–5516 10.1016/j.febslet.2007.10.061 [DOI] [PubMed] [Google Scholar]
- 49. Klose C., Surma M. A., Gerl M. J., Meyenhofer F., Shevchenko A., and Simons K. (2012) Flexibility of a eukaryotic lipidome: insights from yeast lipidomics. PLoS One 7, e35063 10.1371/journal.pone.0035063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Casanovas A., Sprenger R. R., Tarasov K., Ruckerbauer D. E., Hannibal-Bach H. K., Zanghellini J., Jensen O. N., and Ejsing C. S. (2015) Quantitative analysis of proteome and lipidome dynamics reveals functional regulation of global lipid metabolism. Chem. Biol. 22, 412–425 10.1016/j.chembiol.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 51. Donahue T. F., and Henry S. A. (1981) myo-Inositol-1-phosphate synthase: characteristics of the enzyme and identification of its structural gene in yeast. J. Biol. Chem. 256, 7077–7085 [PubMed] [Google Scholar]
- 52. Carman G. M., and Han G. S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem., 80, 859–883 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salsaa M., Case K., and Greenberg M. L. (2017) Orchestrating phospholipid biosynthesis: phosphatidic acid conducts and Opi1p performs. J. Biol. Chem. 292, 18729–18730 10.1074/jbc.H117.809970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoppen J., Repenning A., Albrecht A., Geburtig S., and Schüller H. J. (2005) Comparative analysis of promoter regions containing binding sites of the heterodimeric transcription factor Ino2/Ino4 involved in yeast phospholipid biosynthesis. Yeast 22, 601–613 10.1002/yea.1209 [DOI] [PubMed] [Google Scholar]
- 55. Robinson K. A., and Lopes J. M. (2000) Survey and summary: Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28, 1499–1505 10.1093/nar/28.7.1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reimand J., Vaquerizas J. M., Todd A. E., Vilo J., and Luscombe N. M. (2010) Comprehensive reanalysis of transcription factor knockout expression data in Saccharomyces cerevisiae reveals many new targets. Nucleic Acids Res. 38, 4768–4777 10.1093/nar/gkq232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu X., and Keeney S. (2015) High-resolution global analysis of the influences of Bas1 and Ino4 transcription factors on meiotic DNA break distributions in Saccharomyces cerevisiae. Genetics 201, 525–542 10.1534/genetics.115.178293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hanscho M., Ruckerbauer D. E., Chauhan N., Hofbauer H. F., Krahulec S., Nidetzky B., Kohlwein S. D., Zanghellini J., and Natter K. (2012) Nutritional requirements of the BY series of Saccharomyces cerevisiae strains for optimum growth. FEMS Yeast Res. 12, 796–808 10.1111/j.1567-1364.2012.00830.x [DOI] [PubMed] [Google Scholar]
- 59. Elble R. (1992) A simple and efficient procedure for transformation of yeasts. BioTechniques 13, 18–20 [PubMed] [Google Scholar]
- 60. Amberg D. C., Burke D. J., and Strathern J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, pp. 109–111, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 61. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 10.1002/(SICI)1097-0061(199807)14:10%3C953::AID-YEA293%3E3.0.CO%3B2-U [DOI] [PubMed] [Google Scholar]
- 62. Surlow B. A., Cooley B. M., Needham P. G., Brodsky J. L., and Patton-Vogt J. (2014) Loss of Ypk1, the yeast homolog to the human serum- and glucocorticoid-induced protein kinase, accelerates phospholipase B1-mediated phosphatidylcholine deacylation. J. Biol. Chem. 289, 31591–31604 10.1074/jbc.M114.581157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schneider B. L., Seufert W., Steiner B., Yang Q. H., and Futcher A. B. (1995) Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11, 1265–1274 10.1002/yea.320111306 [DOI] [PubMed] [Google Scholar]
- 64. Yang K., and Han X. (2011) Accurate quantification of lipid species by electrospray ionization mass spectrometry: meet a key challenge in lipidomics. Metabolites 1, 21–40 10.3390/metabo1010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith C. A., Want E. J., O'Maille G., Abagyan R., and Siuzdak G. (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- 66. De Smet C. H., Vittone E., Scherer M., Houweling M., Liebisch G., Brouwers J. F., and de Kroon A. I. (2012) The yeast acyltransferase Sct1p regulates fatty acid desaturation by competing with the desaturase Ole1p. Mol. Biol. Cell 23, 1146–1156 10.1091/mbc.e11-07-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seideman J. G., Smith J. A., and Struhl K. (1999) Short Protocols in Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- 68. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 70. Volland C., Urban-Grimal D., Géraud G., and Haguenauer-Tsapis R. (1994) Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem. 269, 9833–9841 [PubMed] [Google Scholar]