Figure 3.
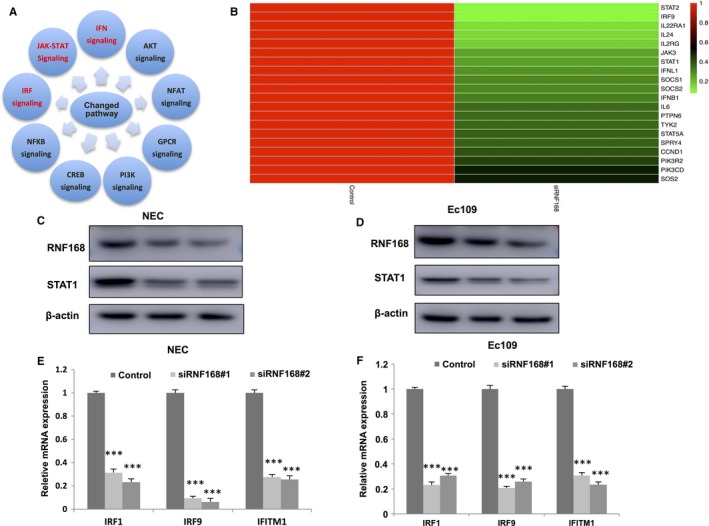
RNF168 depletion decreases STAT1 protein level and JAK‐STAT target genes in esophageal cancer cells. (A) Top 10 signalling pathways significantly decreased by RNF168 depletion in NEC cells. The pathway‐enrichment analysis was used by the threshold P < 0.001 and fold change >2 to derive regulated genes. RNF168 was depleted by siRNA (mix of siRNF168 #1 and siRNF168 #2) or treated with siControl. After 48 h, the whole mRNA was extracted for RNA sequence analysis. The siControl and siRNF168 were done in triplicates. B: The heat‐map graph shows the JAK‐STAT target genes, which is significantly decreased by RNF168 depletion in NEC cells. The significantly regulated genes were overlapped with publish JAK‐STAT target gene data. (C, D) RNF168 depletion effect on STAT1 protein level by two different siRNA oligos. NEC and EC109 cells were transfected with siRNF168 or siControl. After 48 h, RNF168 and STAT1 protein levels were determined by Western blot analysis. Actin was used as internal control. (E, F) RNF168 depletion decreases STAT1 target genes using two different siRNA oligos. NEC and EC109 cells were transfected with siRNF168 or siControl. After 48 h, cells, total RNA was prepared and the expression of the endogenous STAT1 target genes, IRF1, IRF9 and IFITM1 were determined by qPCR. Shown are the results from three experiments. *P < 0.05; **P < 0.01; ***P < 0.001 for target gene expression comparison
