Figure 1.
4,930 SGBs Assembled from 9,428 Meta-analyzed Body-wide Metagenomes
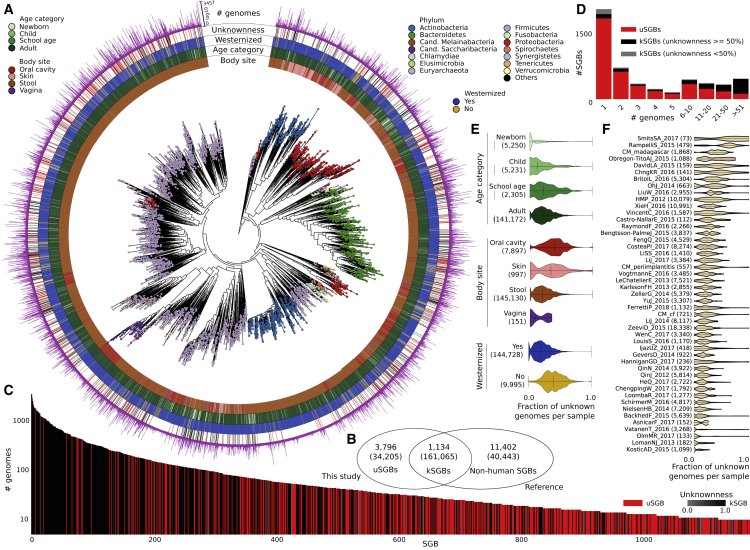
(A) A human-associated microbial phylogeny of representative genomes from each species-level genome bin (SGB). Figure S3A reports the same phylogeny but including isolate genomes not found in the human-associated metagenomes.
(B) Overlap of SGBs containing both existing microbial genomes (including other metagenomic assemblies) and genomes reconstructed here (kSGBs), SGBs with only genomes reconstructed here and without existing isolate or metagenomically assembled genomes (uSGBs), and SGBs with only existing genomes and no genomes from our metagenomic assembly of human microbiomes (non-human SGBs).
(C) Many SGBs contain no genomes from sequenced isolates or publicly available metagenomic assemblies (uSGBs). Only SGBs containing >10 genomes are shown.
(D) Fraction of uSGBs and kSGBs as a function of the size of the SGBs (i.e., number of genomes in the SGB).
(E) Distribution of the fraction of uSGBs in each sample by age category, body site, and lifestyle.
(F) Distribution of the fraction of uSGBs in each study.