Figure 2.
The Expanded Genome Set Substantially Increases the Mappability of Human Metagenomes
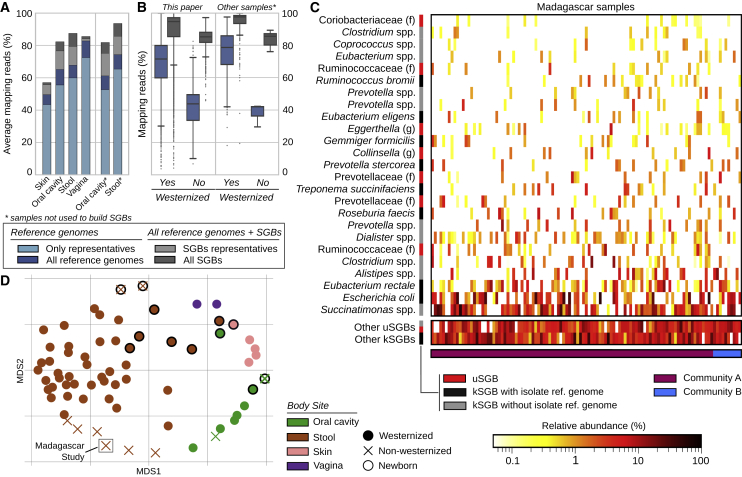
(A) We mapped the subsampled original 9,428 metagenomes and 389 additional samples not considered for building the SGBs against the 154,723 reconstructed genomes and 80,990 previously available genomes. Raw-read mappability increased significantly (Mann-Whitney U test, p < 1e−50), e.g., from an average of 67.76% to 87.51% in the gut. Representative genomes refer to the highest-quality genomes selected from the 4,930 human SGBs and the 11,402 non-human SGBs. Extended statistics are in Figure S4.
(B) Metagenomic read mappability increases more in non-Westernized than Westernized gut microbiomes (Welch's t test, p < 1e−50), both when considering samples used for SGBs’ reconstruction (26.50% average increase in 7,059 Westernized samples versus 96.56% in 454 non-Westernized samples) and when considering 264 additional samples not used for SGBs’ reconstruction (25.16% versus 117.40% average increase, respectively).
(C) The gut microbiomes from Madagascar we sequenced here showed several highly abundant uSGBs and a large set of SGBs reconstructed in only subsets of the samples. Many kSGBs in this dataset do not contain isolate genomes but only previous metagenomic assemblies. The 25 most abundant SGBs are reported and ordered according to their average relative abundance.
(D) Multidimensional scaling on datasets using the Bray-Curtis distance on per-dataset SGB prevalences highlights distinct microbial communities between Westernized and non-Westernized populations within and between body sites and age categories.