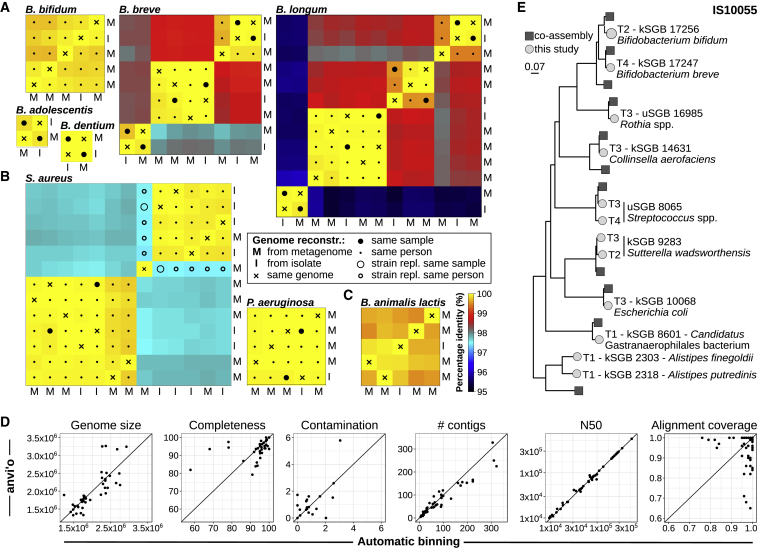
Figure 7.
Quality of the Single-Sample Assembled Genomes against Multiple Alternative Genome Reconstruction Approaches
(A) Percentage identity between genomes from isolates (I) and genomes we reconstructed from metagenomes (M) for five Bifidobacterium species from the FerrettiP_2018 dataset (Ferretti et al., 2018). We mark isolates and metagenomes coming from the same specimen (big filled circles) and coming from specimens of the same mother-infant pair (small filled circles). In all cases, our automatic pipeline reconstructs genomes from metagenomes that are almost identical to the genomes of the expected isolated strains.
(B) The strains of S. aureus and P. aeruginosa isolated from three patients are almost perfectly matching the genomes reconstructed from sputum metagenomes sequenced at multiple time points. In the only case in which a S. aureus genome from a metagenome is not matching the strain isolated from a previous time point in the same patient, we verified with MLST typing that a clinical event of strain-replacement from ST45 to ST273 occurred.
(C) In the dataset by Nielsen et al. (2014), we successfully recover at >99.5% identity the strain of a B. animalis subspecies lactis present in a commercial probiotic product that was consumed by the enrolled subjects, even if the probiotic strain was at low relative abundance in the stool microbiome (<0.3% on average [Nielsen et al., 2014]).
(D) Comparison of the 46 manually curated genomes (using anvi’o) with automatically assembled (using metaSPAdes) and binned (using MetaBAT2) genomes.
(E) Example comparison between the set of single-sample assembled genomes and co-assembled genomes for a time series (n = 5) of gut metagenomes from a newborn. Several genomes reconstructed with the two approaches have the same phylogenetic placement, with single-sample assembly retrieving the same (or a very closely related) genome at multiple time points, and both methods retrieving some unique genomes. This is an example of the comprehensive comparison performed in the STAR Methods and reported in Table S2 and Figure S7B.