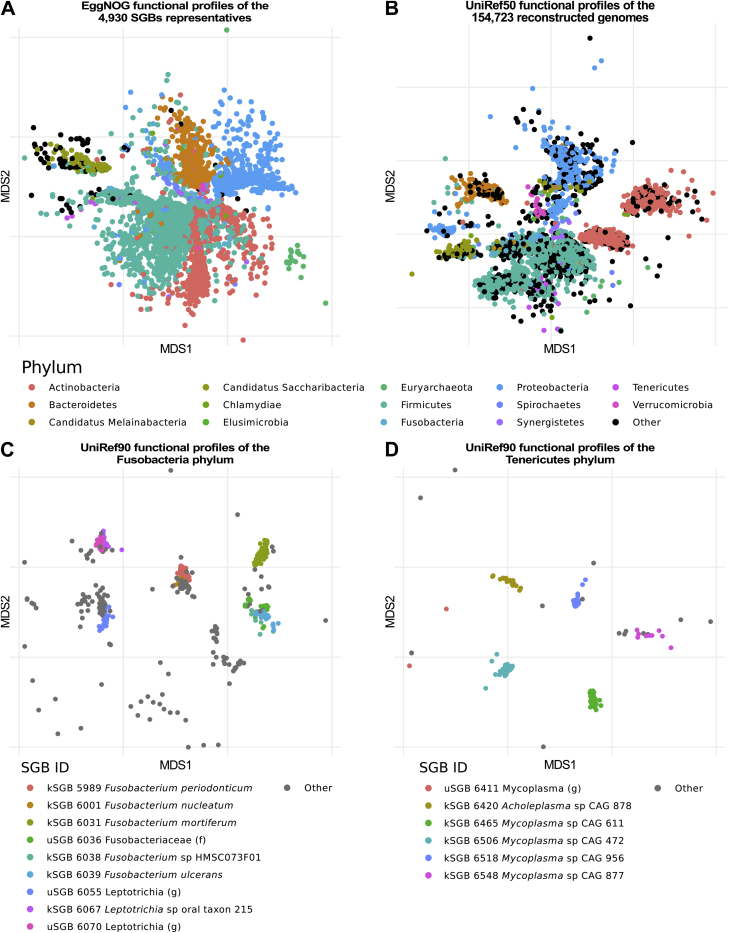
Figure S1.
Overview of the Functional and Metabolic Annotations of the Representatives of the SGBs and of the Whole Set of 154,723 Reconstructed Genomes, Related to Figure 1
(A) Ordination plot of the KEGG gene families annotated using eggNOG (see STAR Methods) of the 4,930 SGBs’ representatives, colored by the 14 most represented phyla. (B) Ordination plot of the UniRef50 gene families present in the 154,723 reconstructed genomes as annotated by mapping the genomes against both Uniref90 and Uniref50 (see STAR Methods). Ordination plots of the UniRef90 gene families for all the reconstructed genomes assigned to the (C) Fusobacteria and (D) Tenericutes phyla are also reported as examples of fine-grained functional differentiation.