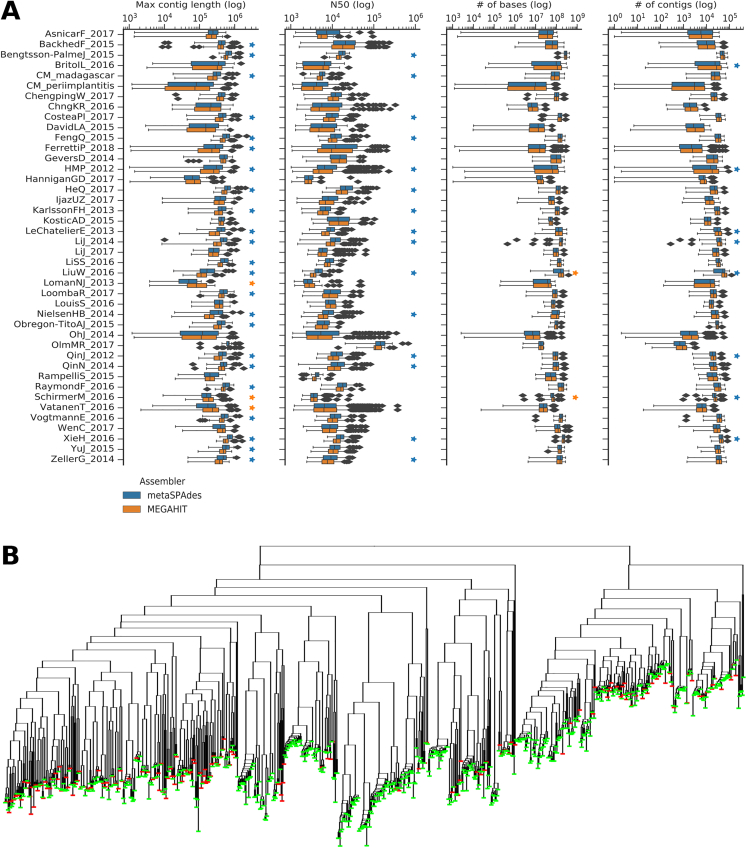
Figure S7.
Comparison between MEGAHIt and metaSPAdes Assemblies and between Assembly and Co-assembly, Related to Figure 7
(A) Comparison between metaSPAdes and MEGAHIT assemblers across all the considered datasets confirms that metaSPAdes performs consistently better especially in recovering long contigs. Stars indicate statistically significance (Welch's t test, p < 0.05). (B) Phylogenetic tree built on the genomes of gut adult metagenomes from 25 women from the FerrettiP_2018 dataset showing comparison between the set of single-sample assembled genomes (in green) and co-assembled genomes (in red). Several genomes reconstructed with the two approaches have the same phylogenetic placement, with single-sample assembly retrieving a total of 605 genomes spanning 257 SGBs, while co-assembly retrieved 172 genomes.