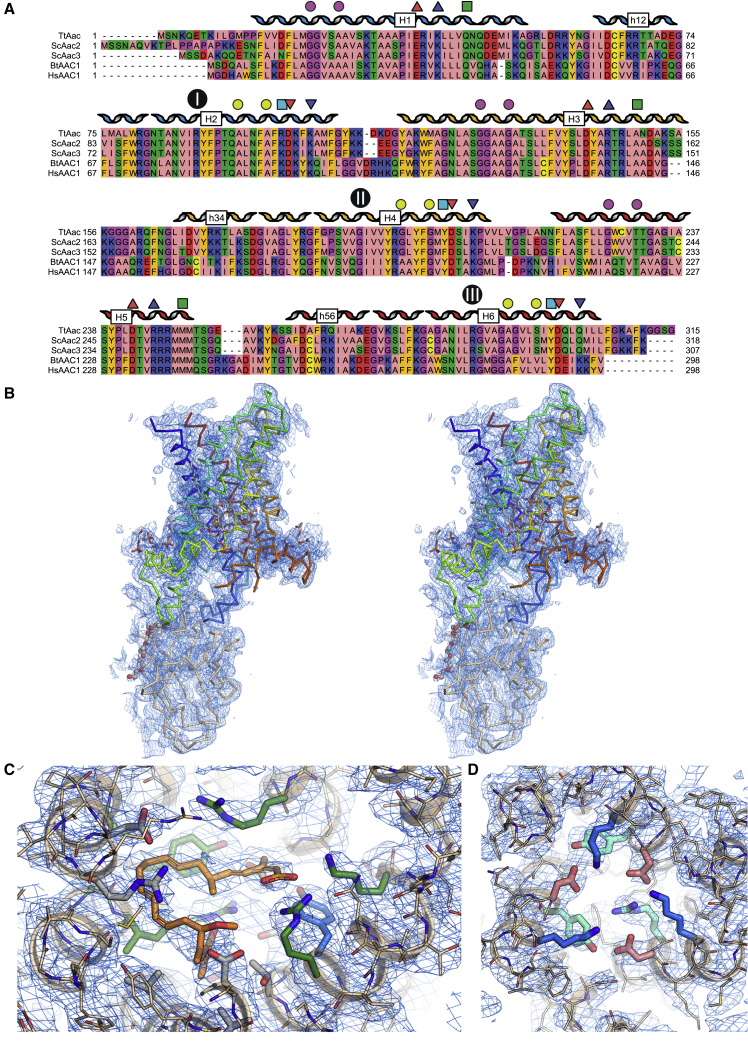
Figure S1.
Alignment of the Amino Acid Sequences of Selected Mitochondrial ADP/ATP Carriers and Representative Electron Density of the TtAac-Nb Complex, Related to Figure 1
(A) Alignment of the mitochondrial ADP/ATP carriers from Thermothelomyces thermophila (TtAac), from Saccharomyces cerevisiae isoform 2 (ScAac2) and isoform 3 (ScAac3), and bovine (BtAAC1) and human (HsAAC1) isoform 1. Amino acids are colored according to their properties: basic K, R and H are blue, acidic D and E are red, polar N, Q, S and T are green, aliphatic A, I, L, M and V are pink, aromatic F, Y and W are orange, structural G and P are magenta, and C is yellow. The negatively charged (red) and positively charged (blue) residues of the matrix and cytoplasmic networks are indicated by up and down triangles, respectively. The positions of the glutamine brace (Q brace) and tyrosine brace (Y brace) are indicated by green and cyan squares, even if they are not conserved in ADP/ATP carriers. The purple and lime circles indicate the positions of the GxxxG and πxxxπ motifs. The contact points of the substrate binding site are shown in black circles with roman numerals (Robinson and Kunji, 2006). (B) Stereo-view showing the contents of the asymmetric unit, with the carrier shown in rainbow colored ribbon representation, and the nanobody as a wheat ribbon. A PEG molecule is shown in ball-and-stick representation, and partially-modeled cardiolipins as sticks. The blue mesh shows the final 2mFo-DFc electron density map, contoured at 1 times the root mean square electron density, and shown within 5 Å of the atoms. (C) Detailed view of the bongkrekic acid-binding site. BKA is shown with orange carbons. Amino acids that are part of the putative substrate-binding pocket are shown with green carbon atoms. Additional amino acids that form salt bridges or hydrogen bonds, or van der Waals interactions, are shown with blue or gray carbon atoms, respectively. The blue mesh shows the final 2mFo-DFc electron density map, sharpened by applying a negative B-factor (B = −45 Å2), contoured at 1 times the root mean square electron density, and shown within 5 Å of the atoms. (D) Detailed view of the cytoplasmic salt bridge network and brace. The blue mesh shows a different view of the map in (C).