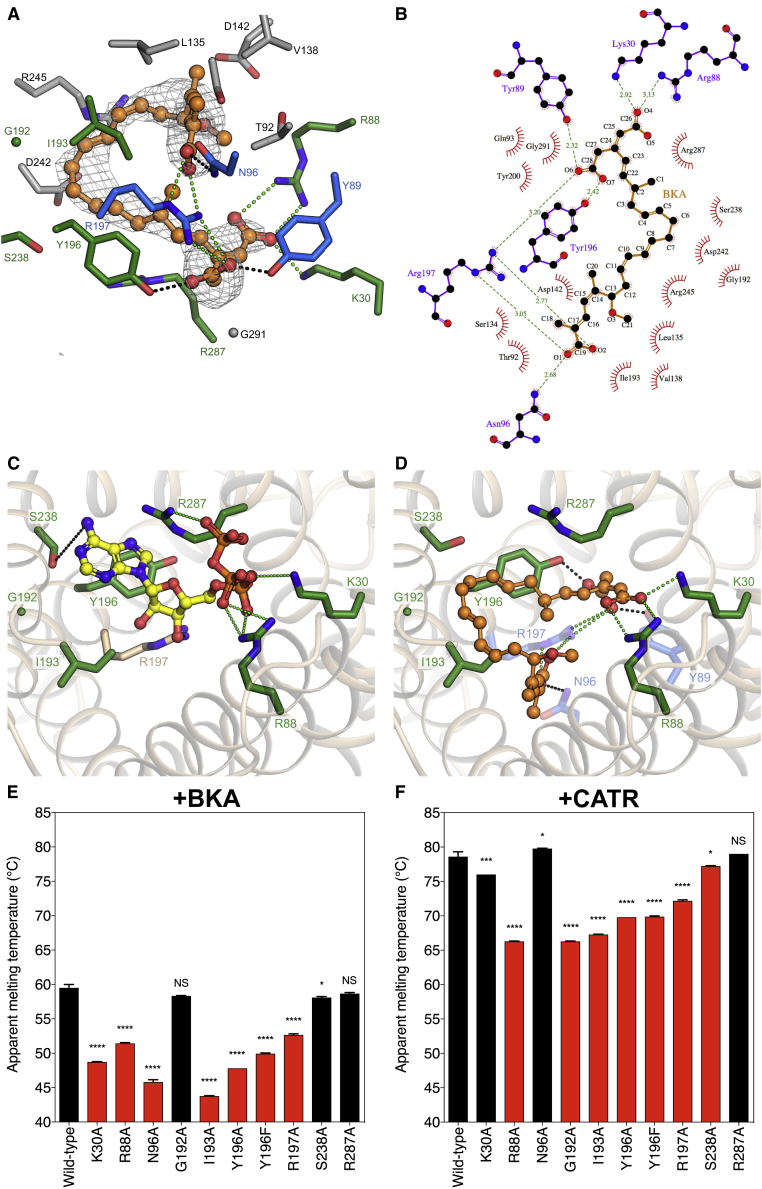
Figure S6.
Binding of Bongkrekic Acid and ATP, Related to Figure 4
(A) mFo-DFc polder OMIT electron density map for BKA, contoured at 3 times the root mean square electron density, shown with the final model. Q93 and Y200, which are in hydrophobic contact with BKA, have been removed to provide a clearer view. (B) Schematic drawing of the binding-site, showing amino acid residues within 4 Å of BKA. Hydrogen bonds and salt bridges are shown as green dashed lines with indicated distances (Å). Red arcs indicate residues in hydrophobic contact with the inhibitor, with spokes radiating toward the atoms they contact. Figure generated by Ligplot+ (Laskowski and Swindells, 2011). (C) Putative binding mode of ATP. The model was generated by docking in AutoDock 4.2 (Forli et al., 2016), treating the protein as a rigid molecule and with S238 modeled as an alternative common rotamer. The model shows the adenine ring of ATP binding to a hydrophobic pocket formed by residues Y196 (aromatic stacking), G192 and I193. S238 may form a hydrogen bond to the adenine ring, and R197 a hydrogen bond to the ribose (black dotted lines). The phosphate groups of ATP interact with the positively-charged residues K30, R88 and R287 (green dotted lines). The features of this binding site mimic those predicted for ADP binding to the c-state (Dehez et al., 2008, Kunji and Robinson, 2006, Mifsud et al., 2013, Robinson and Kunji, 2006, Wang and Tajkhorshid, 2008). (D) View of the BKA binding-site from the matrix side, highlighting residues in the putative substrate-binding site (shown in stick representation). Electrostatic interactions between these residues and BKA are shown as dotted green lines, and hydrogen bonds as dotted black lines. The view is the same as (C) illustrating the overlap between the BKA and ATP-binding site. (E) Mutation of binding-site residues reduces the thermal stability of BKA-inhibited variants. Residues forming polar interactions with BKA, or proximal to the inhibitor, are highlighted in red. (F) Binding-site mutants show increased thermal stability in the presence of CATR, hence are folded correctly and able to bind the inhibitor. CATR induces a greater stabilization than BKA. Mutation of residues that form salt bridges to CATR (R88, R197), or form the binding-pocket (G192, I193, Y196, S238) (Ruprecht et al., 2014), reduces the apparent melting temperature of the CATR-inhibited protein. These residues are shown with red bars. Mutation of residues distal to CATR (K30, S238) results in a smaller reduction in apparent melting temperature. In (E) and (F), the apparent melting temperature is represented by the mean ± S.D., with n = 3. p > 0.05, not-significant (NS); p > 0.01, ∗; p > 0.0001, ∗∗∗; p > 0.00001, ∗∗∗∗ by two-tailed Student’s t test.