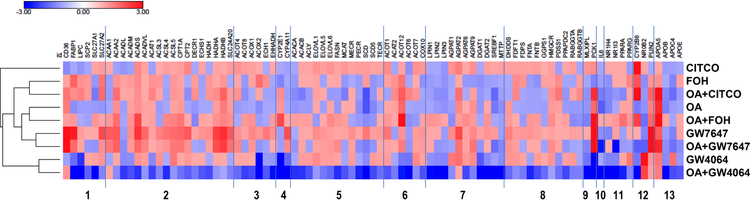
Figure 4. Heat map showing effects of FOH, OA, and nuclear receptor agonists on lipidmetabolizing gene expression.
HepaRG cells were pre-treated for 24 hours in HepaRG treatment medium containing FF-BSA alone or complexed with 0.66 mM OA. The cells were then incubated for 48 hours (media change after 24 hours) in pre-treatment medium (i.e., FFBSA or OA) containing 0.1% EtOH, 100 μM FOH, 0.1% DMSO, 0.1 μM CITCO, 10 μM GW7647, or 1 μM GW4064. Cells were then harvested, and mRNA levels of 96 genes were measured using a customized PCR Array, as described in Materials and Methods. Effects of treatments normalized to their respective vehicle controls (EtOH for farnesol, DMSO for nuclear receptor agonists) are presented as a heat map, where positive log-ratios (log2-fold change) for upregulated genes are shown in red and negative log-ratios for downregulated genes are shown in blue. Treatment groups are hierarchically clustered using one minus Pearson correlation clustering. Functional classifications of genes are (1) fatty acid uptake and transport, (2) mitochondrial fatty acid oxidation, (3) peroxisomal β-oxidation, (4) microsomal ω-oxidation, (5) fatty acid biosynthesis, (6) fatty acid metabolism (other), (7) TG biosynthesis, (8) cholesterol metabolism, (9) carbohydrate metabolism, (10) inflammation, (11) nuclear receptors, (12) nuclear receptor targets, and (13) apolipoproteins.