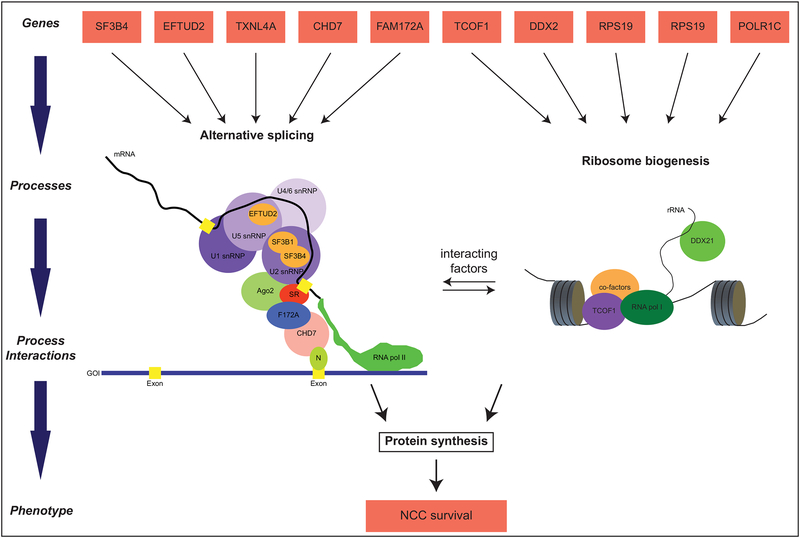
Figure 1: Developmental processes integrating ribosomopathies and spliceosomopathies.
Relationships between genes and traits, shown in red boxes, are modeled to illustrate complexity and show processes integrating ribosomopathies and spliceosomopathies. Alternative splicing (left) and ribosome biogenesis (right) are two connected molecular processes upstream of protein synthesis. The spliceosomal small nuclear ribonucleoproteins (snRNPs), shown in purple, catalyze the splicing of exons, shown in yellow, in nascent mRNA molecules. Splicing factors associated with developmental defects are depicted as orange ovals inside their corresponding snRNPs. Other factors that link transcription and splicing are shown interacting with the spliceosome and RNA polymerase II. In the left panel, RNA polymerase I is shown synthesizing a strand of rRNA while interacting with TCOF1 and other co-factors. Both these molecular processes lead to protein synthesis, which is crucial for neural crest cell survival.