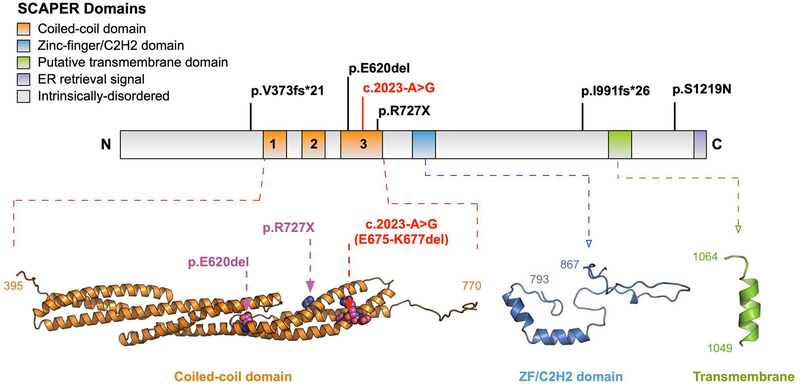
Figure 3. Schematic of the SCAPER protein demonstrating the different domains and the location of all known pathogenic variants identified in SCAPER relative to the putative domains.
The canonical splice variant (c.2023-A>G) marked in red was identified in our patient and has also been previously reported in two siblings. This variant is predicted to delete 3 residues (E675-K677) in the coiled-coil domain. The p.E620del and p.R727X variants are also located in this region. The p.V373fs*21, p.I991fs*26, and p.S1219N variants are located in predicted unstructured regions of the protein. No known variants are in the putative zinc-finger/C2H2 and transmembrane domains.