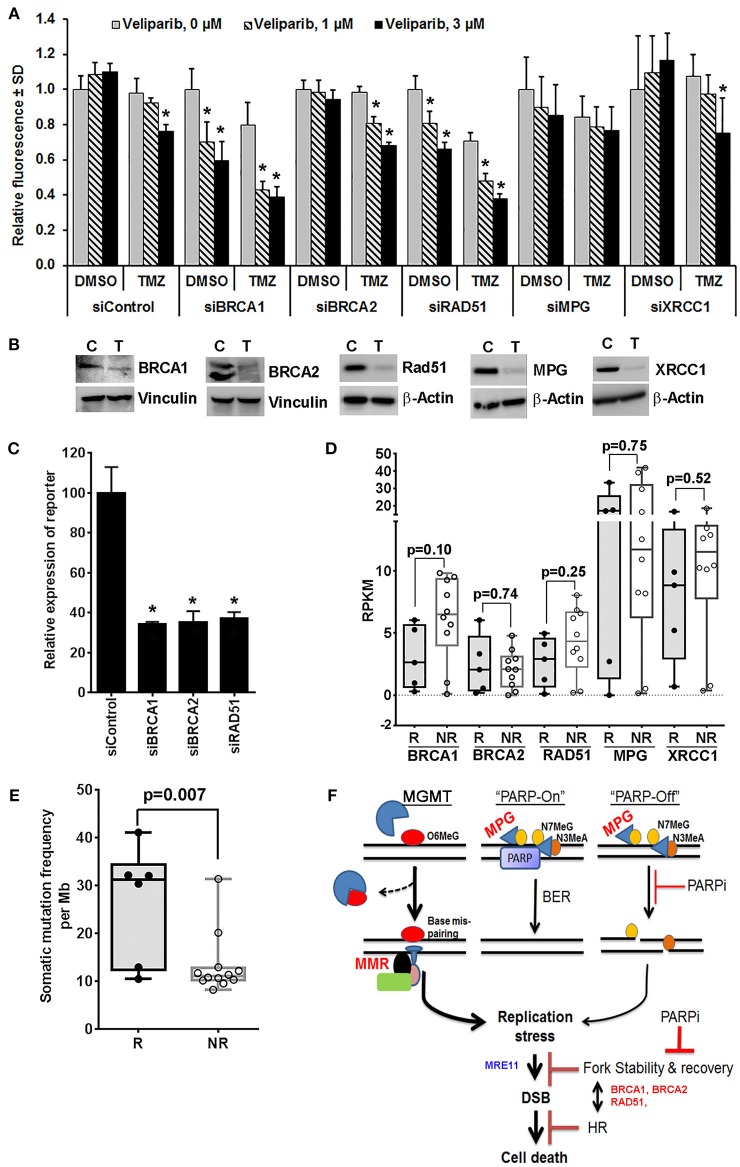
Figure 1.
Mechanistic insights into veliparib-mediated sensitization of TMZ therapy in GBM cells (see Supplementary Material for Materials and Methods). (A) Effect of homologous recombination (HR) vs. base excision repair (BER) pathway disruption on veliparib-mediated sensitization in U251TMZ cells. Cells transfected with specified siRNA were seeded in 96 well plates (500 cells per well), treated with the vehicle or 30 μM TMZ ± 1 or 3 μM veliparib for 5 days and cell growth measured by CyQuant assay. Bar graphs demonstrate change in average fluorescence intensity relative to control, error bars represent standard deviation calculated from 3 replicates in a representative experiment, and *p < 0.05 compared to corresponding control. (B) Western blot analysis to determine level of knockdown for cells used in (A), lanes marked with T represent cells transfected with targeted siRNA and C represent cells transfected with control siRNA. (C) Bar graphs showing effects of BRCA1, BRCA2 or RAD51 knockdown on HR efficiency. U251TMZ-DRGFP cells were transfected with specific siRNA along with plasmid pCBASceI encoding I-SceI restriction enzyme and incubated for 72 h followed by quantification of GFP expressing population by FACS analysis. *p < 0.05 as compared to control. (D) Box plots showing expression levels for specified genes, RKPM values were extrapolated from RNA-Seq data of PDX lines differentially sensitized by veliparib in preclinical PDX trial. Data shown are for 5 of 6 responders (R) lines, which had significantly improved survival vs. 10 of 16 non-responder (NR) lines, which had no significant survival improvement with veliparib/TMZ therapy over TMZ alone in preclinical PDX trial reported previously. two tailed p-values reported were calculated by unpaired t-test. (E) Box plots showing mutation burden based on whole exome seq data available for 21 MGMT methylated PDX lines used in PDX pre-clinical trial and plotted grouped as TMZ/veliparib Responsive (R) vs. Non-responsive (NR) models. SNVs and INDELs across 346 genes involved in DNA damage recognition or repair were analyzed for mutation burden, two tailed p-values reported were calculated by unpaired t-test. (F) Hypothetical model of potential mechanism of the sensitizing effect of PARP inhibition on TMZ therapy in vivo. O6MeG, O6-methylguanine; N7MeG, N7-methylguanine; N3MeA, N3-methyladenine; MPG, methyl purine glycosylase; PARP, poly-ADP-ribose polymerase; PARPi, poly-ADP-ribose polymerase inhibitor; MMR, mismatch repair; DSB, double strand DNA breaks; MRE11, Meiotic Recombination 11 Homolog; BRCA1 and BRCA2, BReast CAncer genes 1 and 2; and HR, homologous recombination.