Fig. 3.
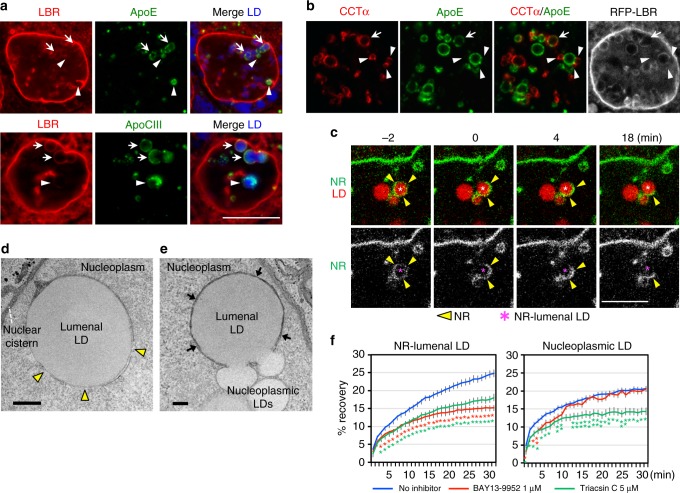
Nucleoplasmic LDs derive from NR-lumenal LDs. a ApoE and ApoCIII (green) were labeled in NR-lumenal LDs (arrows) and in some nucleoplasmic LDs (arrowheads). RFP-LBR (red), LDs (blue). Bar, 10 μm. Huh7 treated with OA/TM for 48 h is shown in this and subsequent figures. b CCTα (red) colocalized with ApoE (green) in the same LD (arrow) or distributed in small nucleoplasmic LDs (arrowheads) binding to ApoE-positive NR-lumenal LDs. The LBR ring is not distinct around the LD showing CCTα–ApoE colocalization. Bar, 10 μm. c Live imaging showed that the complete LBR ring (green) around an NR-lumenal LD (red) disintegrates and releases the LD into the nucleoplasm. Bar, 10 μm. Selected frames from Supplementary Movie 1. d The type I NR membrane around an NR-lumenal LD shows defects (arrowheads). HRP-KDEL was used to delineate the NR lumen by DAB precipitates. Bar, 0.5 μm. e Coalescence between NR-lumenal and nucleoplasmic LDs through defects in the NR membrane. Arrows indicate the NR. Bar, 0.2 μm. See Supplementary Fig. 3b for serial sections. f FRAP of LDs labeled with BODIPY 558/568-C12. Fluorescence recovery in NR-lumenal LDs was retarded by both triacsin C and BAY 13-9952, whereas that in nucleoplasmic LDs was affected only by triacsin C. Nucleoplasmic LDs clearly separated from LBR rings were chosen. The number of LDs examined: 41 (control), 44 (triacsin C), 40 (BAY 13-9952) (NR-lumenal LDs); 36 (control), 36 (triacsin C), 21 (BAY 13-9952) (nucleoplasmic LDs). Mean ± SEM. *p < 0.01, **p < 0.05, one-way ANOVA followed by Tukey test. See also Supplementary Fig. 3c. Source data are provided as a Source data file