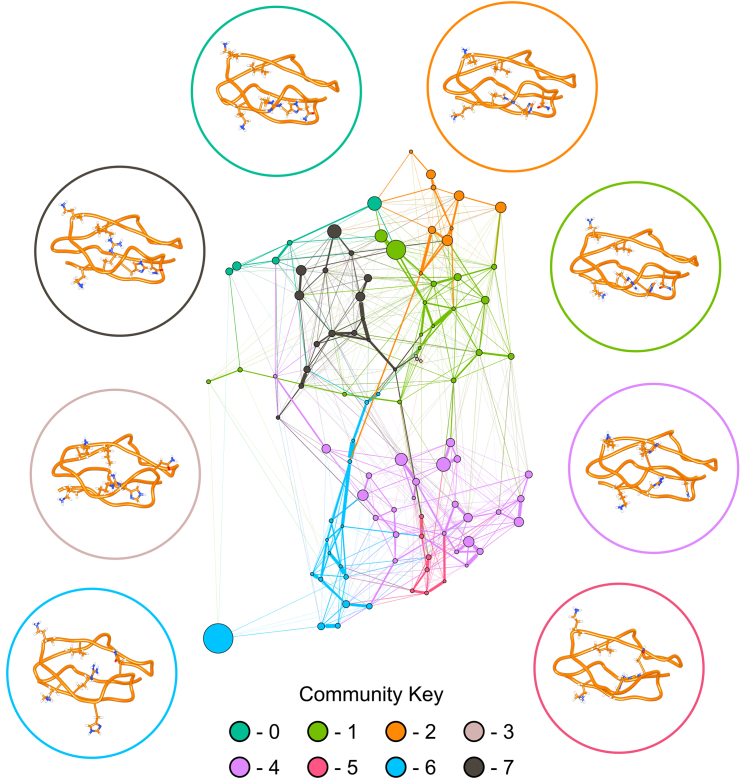
Figure 4.
Graphical representation of conformational sampling of SCR7H402 is shown with representative structures for each community within the network. Nodes are conformational states from the discretized trajectories that are scaled in size according to the associated probability from the stationary distribution of the Markov chain. Edges represent transitions between conformational states and are scaled according to the associated probability from the Markov chain. Nodes are colored according to community membership, and a representative structure of SCR7Y402 is shown for each community. Circles surrounding conformational states denote community membership of each structure, and amino acid side chains for residues N399, H402, R404, K405, K410, and I412 are displayed. Compared to the network for SCR7Y402 (Fig. 3), the network for SCR7H402 appears far less modular because there are more edges between nodes of different communities. To see this figure in color, go online.