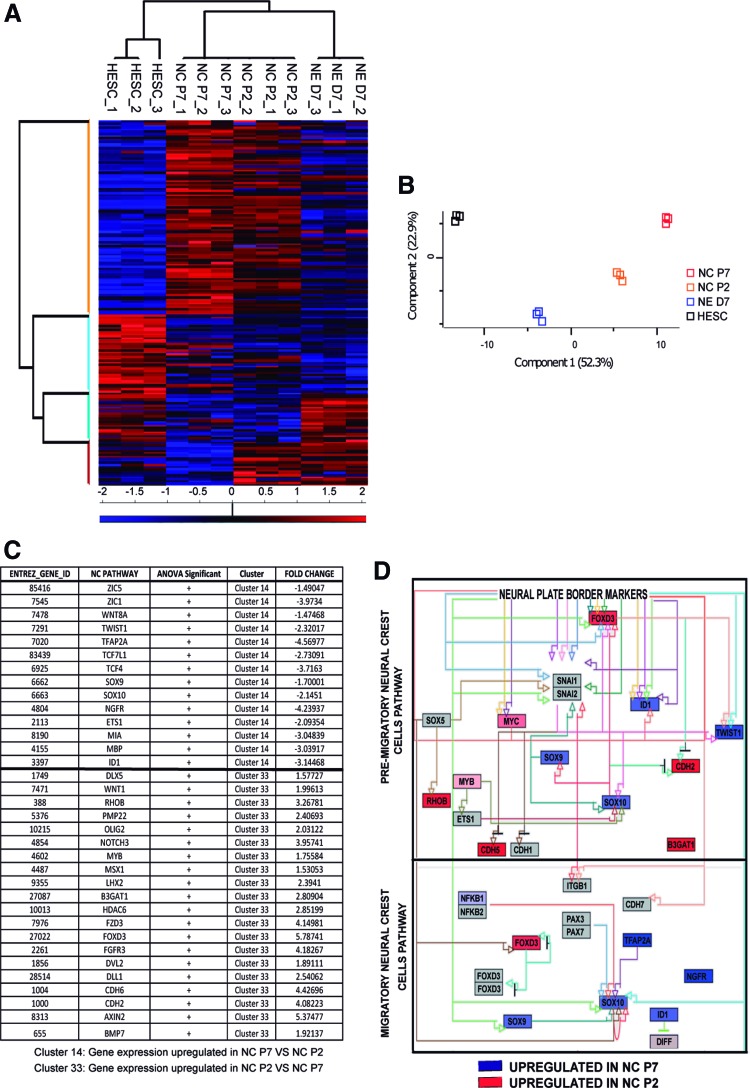
FIG. 3.
NC P2 cells are a mix of premigratory NC and neural progenitor cells that are purified into migratory NC cells with serial passages (P7). (A) Heat map showing the Euclidean distance metric among HESC, NC P2, NC P7, and NE D7 populations. The global gene expression patterns were sorted based on similarity by hierarchical clustering. Red (upregulation) and blue (downregulation) depict differential gene expression from the mean across all samples. NC P2, NC P7: H9s HESC treated 4 days with FSB then split as single cells for two or seven passages, respectively. NE D7: NE cells obtained from H9s HESC after 7 days in FSB. (B) Principal component analysis showed the NC P2 population as an intermediate population between NE and NC P7. (C) Fold change gene expression of NC and neural markers between NC P2 and NC P7 populations. Cluster 14 depicts the NC genes upregulated in NC P7 compared with NC P2. Cluster 33 illustrates the NC genes upregulated in NC P2 compared with NC P7. Benjamini–Hochberg FDR 0.05 was used as a cutoff for ANOVA of differentially expressed genes. (D) Gene interaction pathway for NC cell differentiation (WikiPathway WP2064 revision 47071). Differentially expressed genes are colored in red for upregulated gene expression in NC P2 or blue for upregulated gene expression in NC P7. PathVisio software was used to depict the NC pathway. Legend: Arrows represent stimulatory interaction, while T lines represent inhibition. The coloration intensity of the box is dependent on the level of differential expression between these two populations. Those which are highest in NC P7 are blue, and those highest in NC P2 are red. ANOVA, analysis of variance; FDR, false discovery rate. Color images available online at www.liebertpub.com/scd