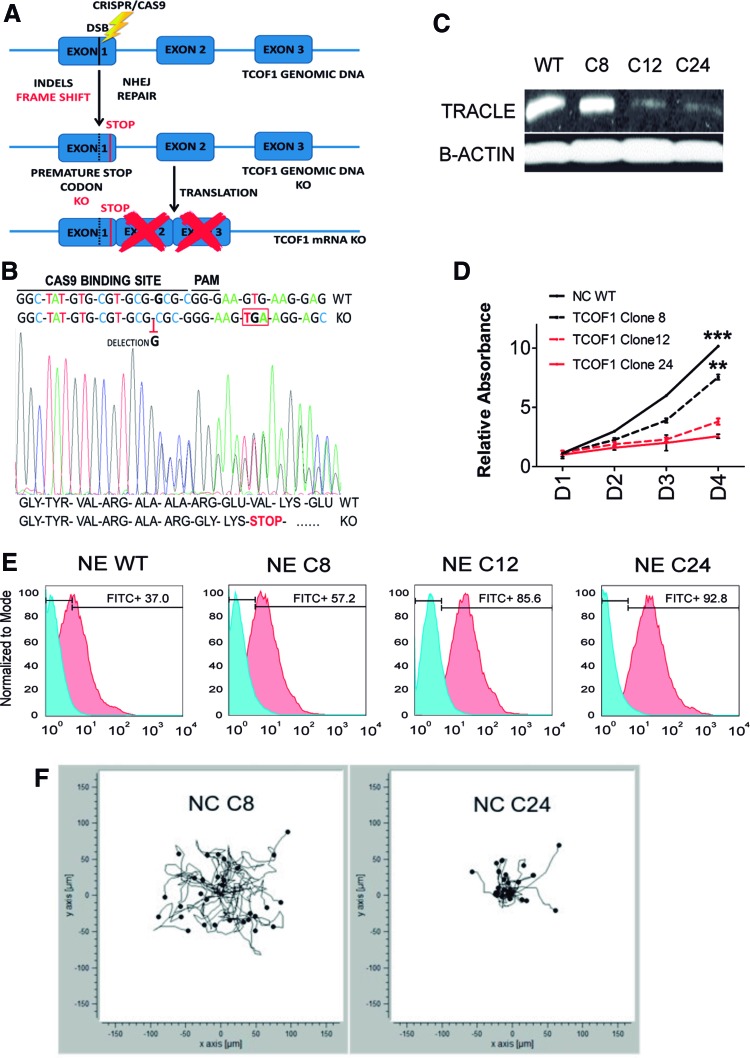
FIG. 7.
Generation of TCOF1 heterozygous knockout HIPSC using CRISPR/Cas9. (A) Schematic of CRISPR/Cas9-mediated nonhomologous end joining strategy to generate INDELs leading to HIPSC TCOF1 knockout clones. (B) Representative genomic sequencing of HIPSC transfected with specific CRISPR TCOF1 gRNA. Heterozygous knockout clones showed the same deletion leading to a change in codon reading frame and the introduction of a premature STOP codon in TCOF1 Exon 1. (C) Immunoblot for Treacle protein demonstrating that NC derived from HIPSC TCOF1+/− clones (C12 and C24) showed a reduction in Treacle compared with NC derived from HIPSC TCOF1+/+ (C8) and NC derived from H9s (WT). (D) Time course MTT proliferation assay of TCOF1+/+ NC derived from H9s (WT), HIPSC (C8), and NC derived from TCOF1+/− HIPSC (C12 and C24), over a 4-day period. Proliferation rate decreased significantly in mutated cells compared with WT. Results are presented as mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001, two-sided Student's t-test. (E) Flow cytometric analysis of Annexin V staining depicting the apoptotic rate in TCOF1+/+ NE-derived cells from H9s (WT), HIPSC (C8), and NE derived from TCOF1+/− HIPSC (C12 and C24). Red histograms represent Annexin V staining, and blue histograms represent the unstained population. (F) Single cell analysis of cell migration in NC derived from TCOF1+/+ HIPSC (C8) and TCOF1+/− HIPSC (C24). TCOF1+/− NC demonstrates impaired migration and reduced directionality of movement. Each dot and tail represent a single cell analyzed, with 30 cells analyzed from both conditions, over a 12-h period. gRNA, guide RNA; HIPSC, human induced pluripotent stem cell; INDELs, insertions or deletions; WT, wild type. Color images available online at www.liebertpub.com/scd