Fig. 1.
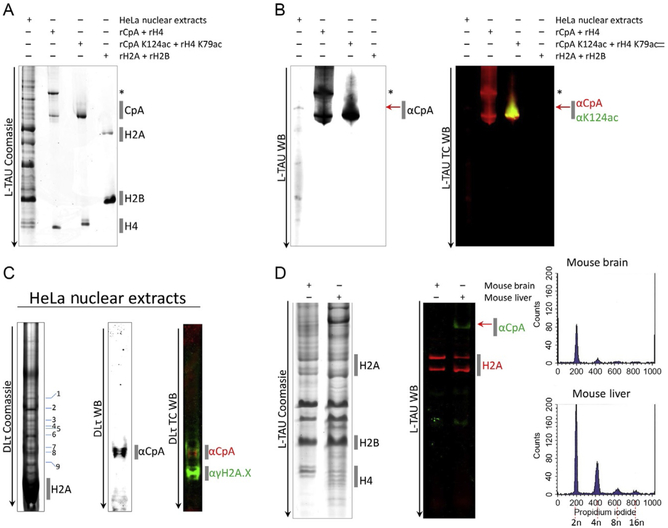
Separating histones from cells and tissues on traditional long Triton acid urea (L-TAU) and Double Long TAU gels (dLτ). (A) Nuclear extracts from HeLa cells were extracted and ran on an L-TAU alongside recombinant (r) histones expressed in bacteria, and the gel was stained with Coomassie Brilliant Blue to reveal CENP-A and other canonical histone bands. (B) Replicate gel was transferred to nitrocellulose for Western detection using antibodies against CENP-A or CENP-A K124ac. (C) Resolving the gel further by running on a double long TAU (dLτ). The majority of the histones ran off the gel, resulting in further separation of the CENP-A band into modified species running above histone H2A and γH2A.X histone bands. (D) Histones from mouse brain and liver were extracted (Dalal, 2003; Stein, Dalal, & Fleury, 2002) and ran on an L-TAU gel and Western probing against histone H2A and mouse CENP-A. Fluorescence-activated cell sorting (FACS) analysis confirmed the difference in ploidy among the two tissues (2n, 4n, 8n, or 16n). “*” Denotes CENP-A with a 6His tag and red arrow denotes native/endogenous CENP-A. TC, two-color; WB, Western blot.