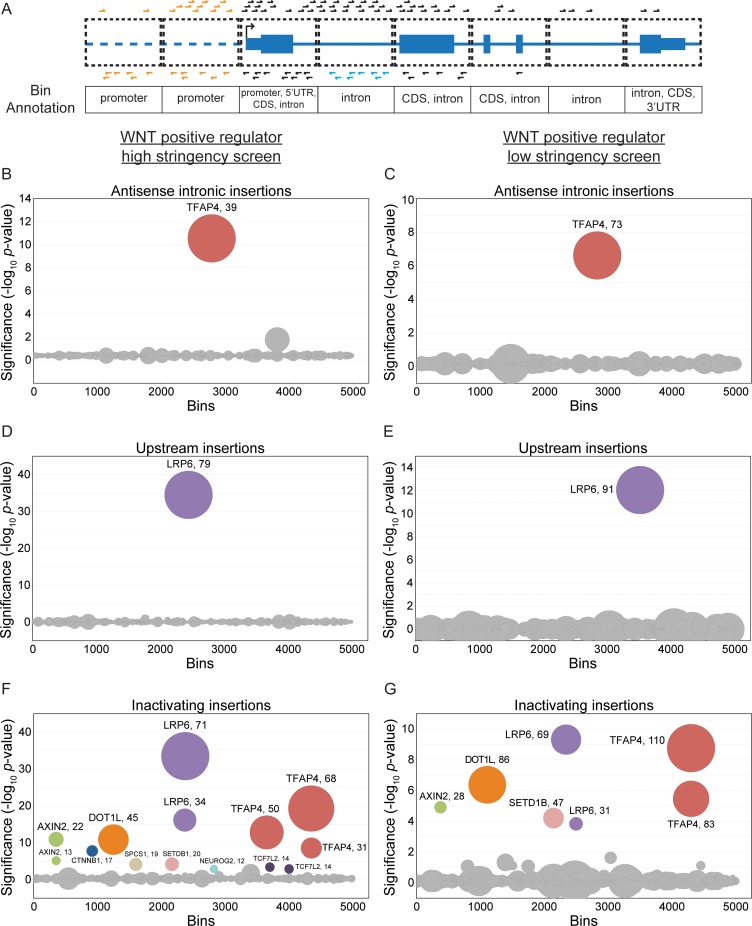
Fig 3. BAIMS identifies atypical GT insertion patterns in screens for regulators of WNT signaling.
(A) Schematic depicting various patterns of GT insertions relative to genetic features in the bins, used for the antisense intronic, upstream, and inactivating insertion enrichment analyses (see text for details). A fictitious gene modeled after a RefSeq gene track, with GT insertions in the sense orientation relative to the gene depicted above the track and in the antisense orientation depicted below it. The antisense intronic insertion enrichment analysis accounts for antisense GT insertions in bins annotated exclusively as intron (depicted in blue) and the upstream insertion enrichment analysis accounts for both sense and antisense insertions in bins annotated exclusively as promoter (depicted in orange). These two classes of insertions had been ignored in previous gene-based analyses of haploid genetic screens [3]. The inactivating insertion enrichment analysis accounts for both sense and antisense insertions in bins annotated as 5’UTR, CDS, or 3’UTR, as well as sense insertions in bins annotated exclusively as intron; these insertions (depicted in black) include all the gene-inactivating insertions used in previous analyses. (B-G) Circle plots depicting the results of antisense intronic (B, C), upstream (D, E), and inactivating (F, G) insertion enrichment analyses for the WNT positive regulator high stringency (B, D, and F) and low stringency (C, E, and G) screens. Circles represent individual 1000 bp bins. The y-axis indicates the significance of GT insertion enrichment in the selected versus the control cells, expressed in units of -log10(FDR-corrected p-value), and the x-axis indicates the 5000 bins with the smallest FDR-corrected p-values, arranged in random order. Circles representing bins with an FDR-corrected p-value < 0.01 are colored and labeled with the name of the gene with which the bin overlaps. Circles representing bins corresponding to the same gene are depicted in the same color. The diameter of each circle is proportional to the number of independent GT insertions mapped to the corresponding bin in the selected cells, which is also indicated next to the gene name for enriched bins.