Figure 3.
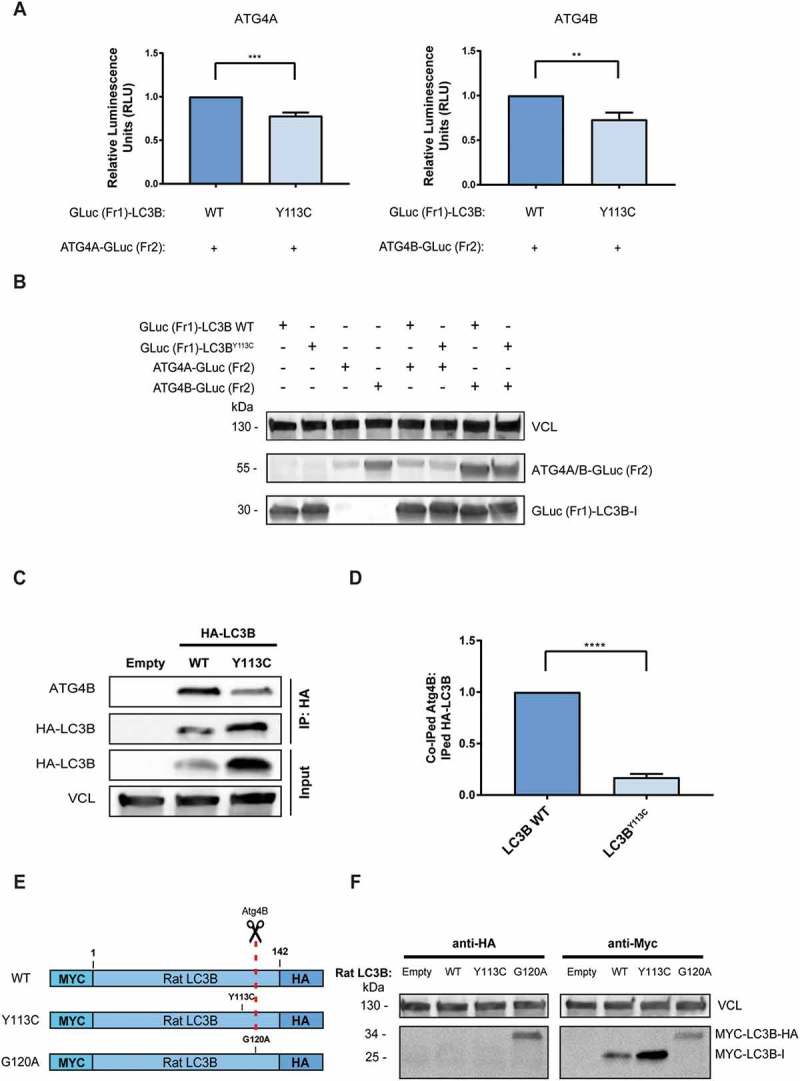
LC3BY113C mutation reduces binding to ATG4B yet does not impair its maturation by cleavage. (a) The relative interaction of GLuc (Fr1)-LC3B with ATG4A-GLuc (Fr2) and ATG4B-GLuc (Fr2) measured by PCA. The calculated results are the mean of 3 independent experiments and normalized to GLuc (Fr1)-LC3B WT. Statistical significance determined by the two-tailed unpaired Student t test, **P < 0.01; ***P < 0.001 (b) Western blot of the PCA samples, reacted with anti-Gaussia antibodies. VCL was used as a loading control. (C) Co-immunoprecipitation of HA-tagged LC3B variants with endogenous ATG4B. VCL was used as loading control for total cell extract. (d) The relative coIPed levels of endogenous ATG4B with LC3B variants (LC3B WT and LC3BY113C). IP levels were quantified using densitometry, and coIPed endogenous ATG4B levels were normalized to IPed HA-LC3B variants. The results are the mean of 3 independent experiments and normalized to the HA-LC3B WT variant. Statistical significance determined by the two-tailed unpaired Student t test, using the Holm-Sidak correction, ****P < 0.0001. (e) Schema of Rat MYC-LC3B-HA constructs used for the LC3B cleavage assay. Red dashed line indicates Gly120 cleavage site by ATG4B protease. (f) HEK293T cells transfected with MYC-LC3B-HA variants (LC3B WT, LC3BY113C, or LC3BG120A were subjected to western blotting with anti-MYC and anti-HA antibodies. Vinculin was used as a loading control.
