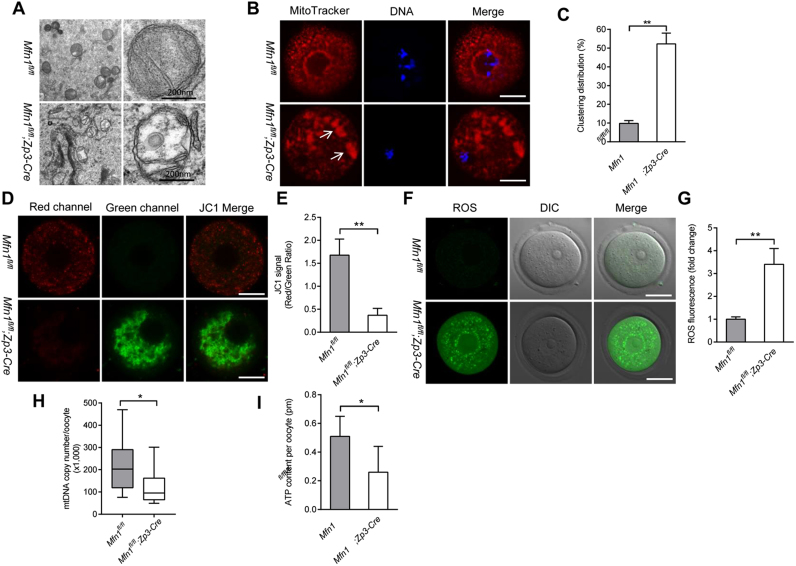
Fig. 5.
Mitochondrial dysfunction in oocytes from Mfn1fl/fl;Zp3-Cre mice. (A) Representative electron micrographs of mitochondria from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes. (B) GV oocytes collected from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice were labeled with MitoTracker Red to visualize mitochondrial localization. (C) Quantification of the proportion of oocytes with clustering mitochondria distribution (n = 35 for each group). (D) Mitochondrial membrane potential in Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes was assessed by JC-1 staining. The green fluorescence shows the inactive mitochondria and the red fluorescence shows the active mitochondria in oocytes. (E) Histogram showing the JC-1 red/green fluorescence ratio (n = 20 for each group). (F) Representative images of CM-H2DCFAD (green) fluorescence in Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre oocytes. (G) Quantification of the relative ROS level (n = 20 for each group). (H) Quantitative analysis of the mtDNA copy number in single oocytes from Mfn1fl/fl and Mfn1fl/fl;Zp3-Cre mice (n = 28 for each group). For each box, the central bar represents the mean; upper and lower boundaries of boxes represent ± SD, and vertical lines extend to the maximal and minimal values. (I) The graph showing the ATP content per oocyte from Mfn1fl/flandMfn1fl/fl;Zp3-Cre mice (n = 30 for each group). Data are expressed as the mean ± SD from three independent experiments. *P< 0.05, **P< 0.01. Scale bars, 30 µm.