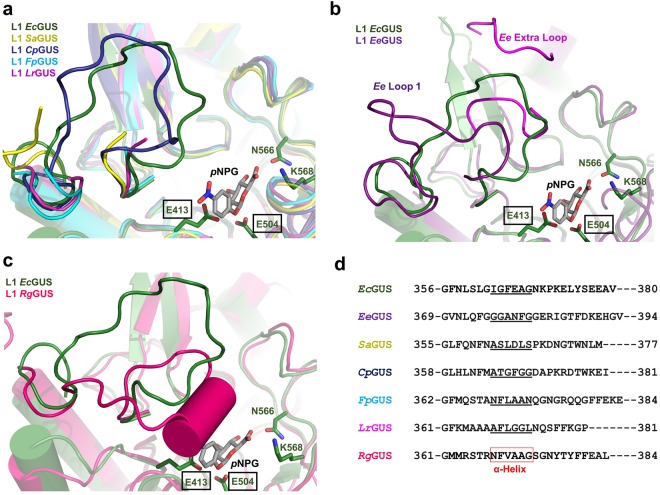
Figure 4.
Alignment of the Loop 1 regions in L1 GUS enzymes. (a) Overlay of EcGUS (3LPG; dark green) with SaGUS (4JKK; yellow), CpGUS (4JKM; indigo), FpGUS (cyan), and LrGUS (magenta), with pNPG shown docked into the active site of EcGUS. (b) Overlay of EcGUS and EeGUS (purple). The canonical Loop 1 is shown in dark purple. The additional active site proximal loop, termed “Ee Extra Loop”, is shown in magenta. Residues G152 through G156 were not previously built and are not shown. (c) Overlay of EcGUS and RgGUS (dark pink) (d) Sequence alignment of the Loop 1 regions from the seven L1 GUS enzymes characterized in this study. Amino acids that form the alpha helix in the Loop 1 of RgGUS are boxed in red. Amino acids that correspond to the alpha helix position in the Loop 1 of RgGUS are underlined.