Abstract
Development of maternal blood transcriptomic markers to monitor placental function and risk of obstetrical complications throughout pregnancy requires accurate quantification of gene expression. Herein, we benchmark three state-of-the-art expression profiling techniques to assess in maternal circulation the expression of cell type-specific gene sets previously discovered by single-cell genomics studies of the placenta. We compared Affymetrix Human Transcriptome Arrays, Illumina RNA-Seq, and sequencing-based targeted expression profiling (DriverMapTM) to assess transcriptomic changes with gestational age and labor status at term, and tested 86 candidate genes by qRT-PCR. DriverMap identified twice as many significant genes (q < 0.1) than RNA-Seq and five times more than microarrays. The gap in the number of significant genes remained when testing only protein-coding genes detected by all platforms. qRT-PCR validation statistics (PPV and AUC) were high and similar among platforms, yet dynamic ranges were higher for sequencing based platforms than microarrays. DriverMap provided the strongest evidence for the association of B-cell and T-cell gene signatures with gestational age, while the T-cell expression was increased with spontaneous labor at term according to all three platforms. We concluded that sequencing-based techniques are more suitable to quantify whole-blood gene expression compared to microarrays, as they have an expanded dynamic range and identify more true positives. Targeted expression profiling achieved higher coverage of protein-coding genes with fewer total sequenced reads, and it is especially suited to track cell type-specific signatures discovered in the placenta. The T-cell gene expression signature was increased in women who underwent spontaneous labor at term, mimicking immunological processes at the maternal-fetal interface and placenta.
Introduction
Human blood is a rich source of molecular information that can be used to develop non-invasive liquid biopsies for specific tumors1 and organs (e.g., the placenta)2 in order to predict disease, its progression, and response to treatments. Genome-wide transcriptomic profiling is particularly well-suited for the discovery of molecular markers3 given the availability of techniques such as microarrays4 and RNA sequencing (RNA-Seq)5,6, which allow simultaneous measurement of tens of thousands of protein-coding and non-coding genes in a given sample.
Cellular and cell-free RNAs in blood that originate from (or are specific to) the primary tumor or organ of interest are especially sought as candidate biomarkers2,7,8 and, more recently, owing to advances in single-cell genomics9, researchers developed cell type-specific signatures of tissues, e.g., the placenta. This approach holds the promise to unravel the complexity of the maternal-fetal molecular dialogue2 and to aid in developing liquid biopsies for prediction of the ‘great obstetrical syndromes’10. Since organ and/or cell type-specific transcripts are expected to have low expression in whole blood, it is essential that quantification of RNA abundance is accurate enough so that modest, and eventually coordinated, gene expression changes can be leveraged as biomarkers that have clinical utility.
The complexity of quantifying low-abundance RNAs using conventional microarrays and RNA-Seq is compounded by the presence of high and variable levels of globin mRNA and ribosomal RNA (rRNA). Although rRNA depletion and globin reduction have been shown to mitigate some of these issues, they require a large amount of total RNA and may induce biases in the quantification of gene expression11. To address these limitations, targeted expression profiling methods were developed based on multiplex RT-PCR amplification followed by quantitative analysis of mRNA abundance by next-generation sequencing. Although microarrays12–15 and RNA-Seq6,16,17 were previously benchmarked for differential expression and prediction model development, preprocessing methods for RNA-Seq have continued to evolve17,18, and direct comparisons to targeted expression profiling by RNA-Seq for genome-wide transcription are not available.
Therefore, the goal of this work was to compare Affymetrix Human Transcriptome Arrays (HTA 2.0) that probe the transcriptome at the exon level, paired-end Illumina RNA-Seq with globin reduction, and a novel genome-wide targeted expression profiling technique DriverMap (https://www.cellecta.com). The comparison was made in terms of ability to identify true maternal whole blood expression changes with gestational age and with the onset of labor at term. Furthermore, we have for the first time evaluated the ability of these high-throughput methods to quantify in maternal whole blood the expression of cell type-specific signatures derived from single-cell genomics of the placenta, and we also have determined whether these signatures are indicative of the onset of term parturition.
Results
RNA was extracted from 32 maternal whole blood samples from women with a normal pregnancy with (n = 8) and without (n = 8) spontaneous labor at term. The two groups will be further referred to as term in labor (TIL) and term not in labor (TNL). One-half of the women in each group had three longitudinal samples taken at 12 to 40 weeks of gestation, while the other one-half of the women had one sample taken at term before delivery (see Table S1). The RNA integrity number (RIN) was very similar among samples (range 6.2 to 7.4) and did not change with sample storage duration (p = 0.6). Data generated from three high-throughput gene expression platforms were made available to the community as a Gene Expression Omnibus super series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113966) and may be valuable in future studies assessing RNA-Seq quantification workflows.
Detection of transcripts in maternal whole blood
Affymetrix Human Transcriptome Arrays (HTA 2.0) probed 30,682 annotated transcript clusters, of which 24,371 coding and 5,389 non-coding transcript clusters were deemed expressed in maternal whole blood. Paired-end Illumina RNA-Seq generated 20.7 to 57.1 million aligned sequence fragments per sample (mean, 39.4 million), enabling detection of 32,880 genes, of which 15,584 were protein-coding genes. When limited only to the 18,559 protein-coding genes targeted by DriverMap, the number of aligned reads per sample obtained for RNA-Seq was 2.6 to 6.6 million (mean, 4.6 million). Targeted profiling of 18,559 protein-coding genes by DriverMap resulted in 11.1 to 12.9 million aligned sequence fragments per sample (mean, 12.0 million), allowing detection of 13,182 genes. Expression profiling for 1/32 samples by DriverMap was not successful due to contamination, and it was not included in downstream analyses.
Differential expression associated with gestation and term labor
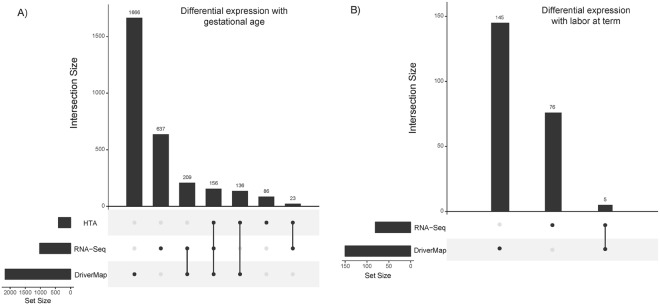
The UpSet plots19 in Fig. 1 summarize the overlap of gene expression changes associated with gestational age (term vs preterm gestation) (Fig. 1A) and with labor at term (Fig. 1B) for the three high-throughput transcriptomic platforms. At the same significance cut-off (false discovery rate20, q < 0.1), 401, 1025, and 2167 genes were found to change with gestation by microarrays, RNA-Seq, and DriverMap, respectively. Of note, 156 differentially expressed genes were identified by all three platforms to change in the same direction, representing 40% of the size of the smallest of the three lists (Fig. 1A). Fewer genes were found to change with labor at term, with only RNA-Seq and DriverMap identifying 81 and 150 genes, respectively, of which only five were in common (GZMB, KLRC1, CD69, and KLRF1 were increased while SPTB was decreased with labor) (Fig. 1B).
Figure 1.
UpSet plots of genes differentially expressed using three transcriptomic platforms. Changes with gestational age (term vs preterm) (left) and with labor status (term in labor vs term not in labor) (right). The significance of gene expression changes was inferred based on an adjusted p-value (q-value) <0.1. The horizontal bars show the number of differentially expressed genes identified by each method, while the vertical bars display the size of sets of genes identified by only one method and the intersection sets.
qRT-PCR validation of gene expression changes
qRT-PCR TaqMan assays (Applied Biosystems, Foster City, CA) were used to profile 86 candidate genes in the same 32 samples, with genes being selected based on evidence for differential expression with either gestational age or labor according to one or more platforms (see Methods). Of note, the average expression over the six house-keeping genes was stable with sample storage duration (linear correlation p = 0.87, Fig. S1). When using qRT-PCR results to define true positive changes (two-tailed moderated t-test, p < 0.05) (see Tables S2 and S3), the validation rate of changes with gestation was 98% (40/41) for microarrays, 94% (33/35) for RNA-Seq, and 88% (44/50) for DriverMap. Of note, although the list of genes included in the calculation of validation rates (i.e. positive predicted values) for each platform represent a different subset of all 86 genes profiled by qRT-PCR, the ranks of validation genes among the list of differentially expressed genes (positive genes) were not significantly different among platforms (ANOVA p = 0.15) (Fig. S2). The validation rate for changes with onset of labor at term was 45% (5/11) for RNA-Seq and 96% (23/24) for DriverMap, and it could not be determined for microarrays given that no gene was significant at q < 0.1. The lower validation rate for RNA-Seq for changes with labor compared to changes with gestation was expected, since for the former condition the genes were selected from the bottom half while in the latter from the upper half of the list of genes ranked by p-values (Figs S2 and S3).
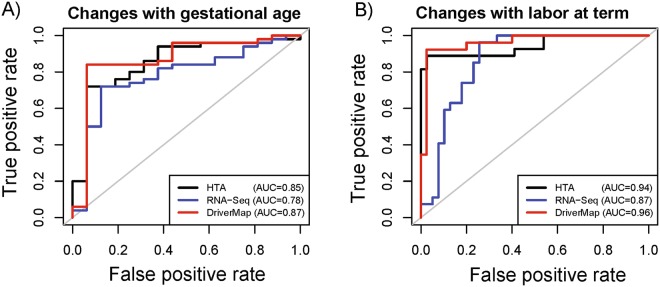
In addition to qRT-PCR validation rates, which can be seen as estimates of positive predicted values (PPV) for each platform (rate of true differentially expressed genes among all genes positives at q-value < 0.1), we have also compared the area under the receiver operating characteristic curve (AUC) among the three platforms based on 66 protein-coding genes. These 66 genes were all of the 86 genes selected for qRT-PCR validation (see Methods section) that were detected present on all three platforms (Fig. 2). To construct the ROC curves for a given platform, the 66 genes were ranked by differential expression p-values obtained with the particular platform. The AUC values for expression changes with gestational age were 0.78, 0.85 and 0.87 for RNA-Seq, HTA, and DriverMAp, respectively. Similarly, for changes with labor at term, the AUC statistics were 0.87, 0.94 and 0.96 for RNA-Seq, HTA, and DriverMap, respectively. Of note, although no gene was differentially expressed with labor based on the HTA platform after multiple testing correction, the ability of this platform to rank truly differentially expressed genes at the top of the list is only slightly lower than the one of DriverMap and surpasses then one of RNA-Seq (Fig. 2B).
Figure 2.
Receiver operating characteristic (ROC) curves for detection of differentially expressed genes. Of the 86 genes profiled by qRT-PCR, 66 were deemed detected by all three platforms and were deemed truly differentially expressed with gestational age (A) and with labor at term (B) if significant by qRT-PCR analysis. HTA: Human Transcriptome Arrays; AUC: area under the curve.
Correlation of expression changes between high-throughput methods and qRT-PCR
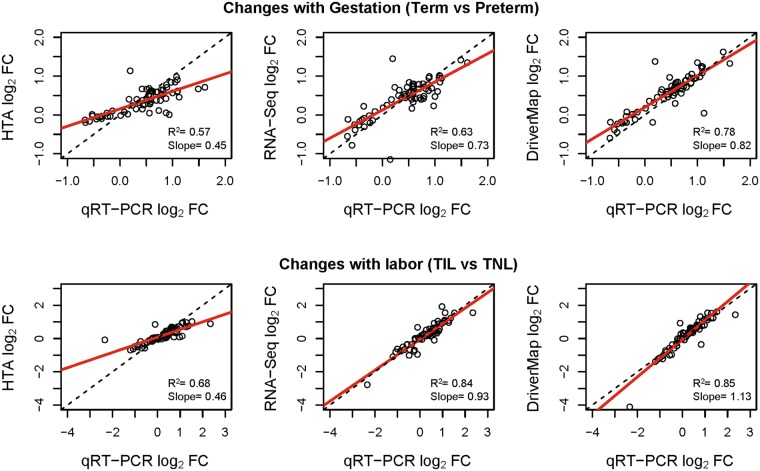
We have conducted a correlation analysis to determine the agreement in the estimated gene expression fold changes (log2 thereof) obtained with high-throughput expression profiling techniques and qRT-PCR. The correlation was based on estimates of log2 fold changes obtained for 66 genes detected present, but not necessarily found significant, with all three platforms. The linear regression slope (95% confidence intervals) between high-throughput methods and qRT-PCR log2 fold-changes was 0.45(0.35–0.55) and 0.46(0.38–0.53) for microarrays, 0.73(0.59–0.86) and 0.93(0.83–1.03) for RNA-Seq, and 0.82(0.71–0.92) and 1.13(1.01–1.25) for DriverMap for changes with gestation (Fig. 3 top) and with labor (Fig. 3 bottom), respectively. Slope estimates were significantly higher for the two sequencing-based methods than for microarrays, yet confidence intervals overlapped between RNA-Seq and Drivermap. In this analysis, slopes <1.0 correspond to a compression, while slopes >1.0 correspond to an expansion of the dynamic range of expression changes compared to qRT-PCR. For instance, the 0.46 slope estimate obtained for microarrays can be interpreted that, in average, a gene showing a 2-fold change in expression by qRT-PCR between TIL and TNL groups, displays a 1.37 fold change with microarrays; however, the corresponding fold changes were 1.93 with RNA-Seq and 2.2 fold with DriverMap (Fig. 3 bottom).
Figure 3.
Correlation of expression changes between high-throughput platforms and qRT-PCR. Of the 86 genes profiled by qRT-PCR, 66 were deemed detected by all three platforms and are displayed as individual dots in this figure. TIL: term in labor; TNL: term not in labor; HTA: Human Transcriptome Arrays; FC: fold change.
Comparison to other differential expression studies
The lists of genes associated with gestational age by each high-throughput platform were overlapped with those reported in previous studies21,22. Heng et al.21 reported paired maternal whole blood gene expression changes from 17–23 weeks to 27–33 weeks of gestation in 114 women with term delivery. The authors used Affymetrix Human Gene 2.1 ST microarrays and found 41 genes up-regulated and four down-regulated at q < 0.05 and fold-change >1.25. To increase the power for testing for an overlap between the list of Heng et al. and this study, we have included all 2,321 genes significant at q < 0.05. Another longitudinal study by Al-Garawi et al.22, using Illumina HumanHT-12 v4 microarrays, reported dramatic changes in the transcriptome from 10–18 weeks to 30–38 weeks of gestation in 30 women included in a trial of vitamin D supplementation. The authors found expression changes associated with gestation in 3,830 unique genes at q < 0.05. To enable a direct comparison between the different transcriptomic platforms, enrichment analyses were limited to genes profiled on all three platforms. We found a significant enrichment in previously reported differential expression associated with gestational age among genes identified by each of the three platforms used herein (odds ratios, OR of 1.4–2.1; hypergeometric test p < 0.01 for all) (Table 1). Enrichment statistics (ORs) were higher for microarrays, yet DriverMap identified a larger number of genes previously reported to change with gestation.
Table 1.
Enrichment analysis of previously reported changes with gestational age among the results of this study.
| Dataset | Platform | Common significant genes (N) | Odds Ratio | p-value |
|---|---|---|---|---|
| Heng et al. | DriverMap | 335 | 1.4 | 2.7E-06 |
| Heng et al. | HTA | 73 | 1.7 | 7.5E-05 |
| Heng et al. | RNA-Seq | 97 | 1.4 | 2.0E-03 |
| Al-Garawi et al. | DriverMap | 711 | 1.8 | 3.5E-31 |
| Al-Garawi et al. | HTA | 142 | 2.1 | 5.8E-11 |
| Al-Garawi et al. | RNA-Seq | 198 | 1.8 | 8.6E-10 |
Quantification of cell type-specific signatures discovered by single-cell genomics
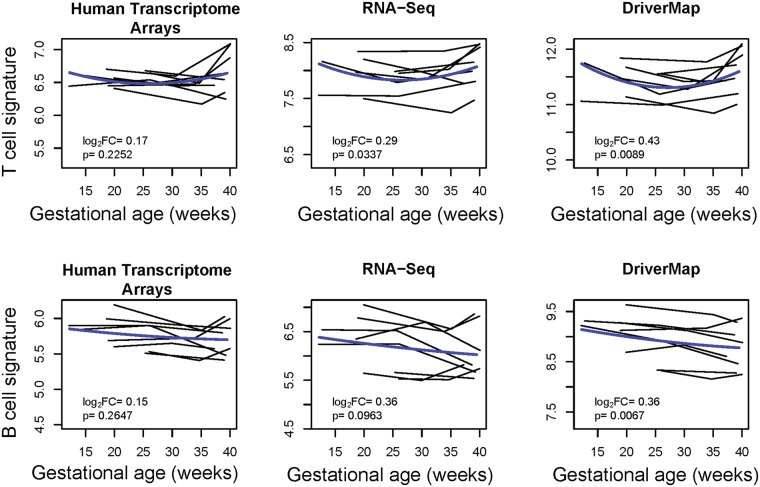
We compared the ability of the three high-throughput platforms to quantify in maternal whole blood the abundance of cell type-specific gene sets discovered by single-cell transcriptomics of the placenta2. In this analysis, 11 cell type-specific gene signatures for which three or more genes were detected by all three platforms were included. According to Tsang et al.2, the B-cell-specific signature decreases monotonically as a function of gestation, while the T-cell-specific signature decreases from the first to second trimester and then increases during the third trimester. Figure 4 shows that, indeed, a significant quadratic u-shaped trend was found by linear mixed-effects models for T-cell expression quantified by RNA-Seq and DriverMap, and a significant linear decreasing trend for B cell expression quantified by DriverMap (all p < 0.05). Although DriverMap data led to stronger associations with gestational age for these signatures (smaller p-values) the confidence intervals of the log2 fold changes overlapped among methods.
Figure 4.
Changes in average expression of cell type-specific genes with gestational age. Gene expression for 17 T-cell-specific genes (top) and 12 B-cell-specific genes were averaged and displayed (y-axis) as a function of gestational age at sampling (x-axis). For microarrays, averages are over log2 normalized expression intensity. For sequencing based techniques, the average is over log2 DESeq2 normalized count data. Each line corresponds to one woman. The blue line represents a linear mixed-effect model fitted by using quadratic splines with one knot.
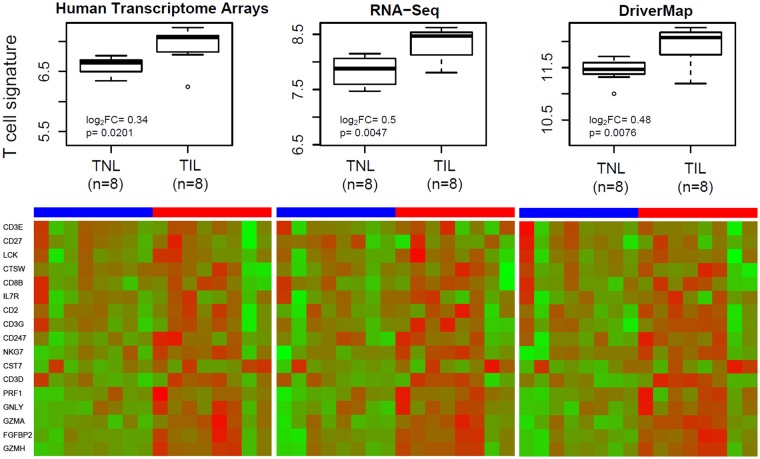
When the cell type-specific gene signatures were tested for the first time for association with the onset of labor at term, we found that the average expression of genes in the T-cell signature, comprised of 17 genes, was higher in women who were in labor compared to those who were not in labor at the time of the blood draw (Fig. 5). The fold-change estimates were higher for RNA-Seq and DriverMap compared to microarrays, yet 95% confidence intervals overlapped [log2 fold change 0.34(0.08, 0.60) for microarrays, 0.5(0.18, 0.81) for RNA-Seq and 0.48(0.16, 0.81) for DriverMap]. The increase of T-cell-specific gene expression with labor at term was also confirmed by qRT-PCR profiling of 4/17 genes (GZMH, GNLY, FGFBP2, and GZMA) (p = 0.0048, 2.5 fold-change).
Figure 5.
Changes of T-cell-specific gene signature with labor status in maternal whole blood. Gene expression for 17 T-cell-specific genes was summarized in each sample collected at term from women in labor (TIL) and not in labor (TNL). Expression levels of individual genes are also shown using a heatmap.
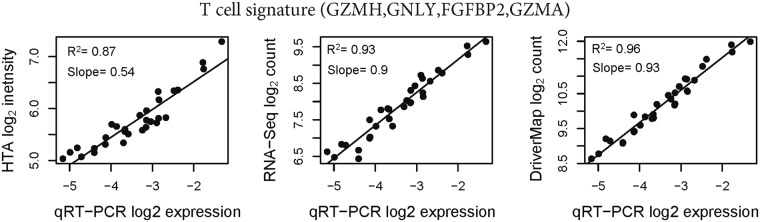
When expression of the T-cell signature was summarized based on four genes (GZMH, GNLY, FGFBP2, and GZMA) for which qRT-PCR data were also available, the correlation with qRT-PCR expression and dynamic range of the T-cell signature average expression across samples were slightly higher for DriverMap than RNA-Seq, and both sequencing-based methods performed better than microarrays (Fig. 6).
Figure 6.
Correlation analysis of T-cell-specific gene signatures between high-throughput methods and qRT-PCR. The x-axis shows qRT-PCR expression averages (−ΔCt) over GZMH, GNLY, FGFBP2, and GZMA genes in individual samples, while the y-axis shows the same summaries derived from high-throughput expression profiling methods.
Discussion
Detection of mRNAs in maternal whole blood
The number of protein-coding genes detected as present in maternal whole blood samples was 24,371, 15,584, and 13,182 for Human Transcriptome Arrays, RNA-Seq, and targeted expression profiling by DriverMap, respectively. Differences in the number of genes detected by microarrays and sequencing techniques in the same samples are to be expected due to platform design differences, experimental differences, level of background noise, and criteria used to determine expression above background16,23,24. While using detection p-values to infer expression above background for microarrays has been widely adopted25,26, a minimum number of aligned sequence fragments is typically required for RNA-seq (e.g., ≥1027, ≥2028), with the stringency of these cut-offs being dependent on the sequencing depth. Our choice of a minimum transcript count of 5 to call a gene detected in a given sample, for which an average 40 million aligned reads were obtained (5/40 = 0.125 transcripts per million [TPM]) is similar to the 0.1 TPM cut-off used in a recent benchmark of RNA-sequencing analysis workflows24. Another important factor contributing to the differences in the number of genes detected by sequencing methods is the depth of coverage29. Although over three times more total aligned reads per sample were obtained for RNA-Seq than for DriverMap, the number of aligned reads per sample for protein-coding gene content was about three times less for RNA-Seq and it varied more across samples compared to DriverMap. Of note, both the HTA microarrays and RNA-Seq probed also non-coding RNAs, and small RNAs that have compelling biological and disease roles30, including in pregnancy31–34.
Gene expression changes associated with gestation and with labor at term
Longitudinal transcriptomic changes associated with gestational age (term vs preterm gestation), and cross-sectional changes associated with labor at term (term in labor vs term not in labor), were assessed using commonly used analytical approaches that borrow information across genes to derive more reliable expression variance estimates35,36. HTA microarrays identified 2.6 times fewer differentially expressed genes than RNA-Seq and 5.4 times fewer than DriverMap at the same false discovery rate (q < 0.1). When a more conservative false discovery rate cut-off was used (q < 0.05), HTA microarrays identified 3.7 times fewer differentially expressed genes than RNA-Seq and 8.6 times fewer than DriverMap (Fig. S4). Although differences in the type of data (continuous intensity for microarray vs read counts for sequencing methods) and, hence, analysis models may be a factor, the higher background noise37,38 resulting in compressed fold-changes with microarray data (Fig. 3) and the more stringent correction required to maintain the same false discovery rate explain, in part, these results. Indeed, when only the genes deemed to be expressed on all three platforms were tested, the number of genes differentially expressed (q < 0.1) with gestation by HTA increased from 401 to 636, narrowing the gap with RNA-Seq while still being more than three times lower than the one for DriverMap (Fig. S5). Our finding that Illumina RNA-Seq identifies more differentially expressed genes compared to microarrays is in line with previous reports39, although those studies used 3′-end biased microarrays as opposed to arrays that probe all exons of the genes, as it was the case herein.
qRT-PCR validation of gene expression changes
Although profiling by DriverMap identified more differentially expressed genes than microarrays and RNA-Seq, the qRT-PCR validation rate (i.e., the fraction of differentially expressed genes confirmed by qRT-PCR among all differentially expressed genes that were tested) was about the same for this platform (92% overall for changes with gestation and with labor) compared to the other two platforms (98% for microarrays and 94% for RNA-Seq for changes with gestation). Of note, for changes with labor, the validation rate of microarrays could not be assessed given that no positive genes were found, while the lower validation rate (45%) for RNA-Seq for changes with labor compared to the one for changes with gestational age was expected due to differences in the ranks of genes tested by qRT-PCR among those differentially expressed (see Figs S2 and S3). It is important that the ranks of genes tested are similar when comparing validation rates among platforms, as one would expect that genes with the smallest p-values (higher ranks) are more likely to be truly differentially expressed than those that appear lower on the list; indeed, this was the case for validation of RNA-Seq results.
While the comparison of validation rates (or PPV) required the use of a significance cut-off to define a positive result (i.e. q < 0.1), the threshold free AUC statistics revealed that the ranking of genes based on nominal p-values is similarly meaningful for microarrays and DriverMap platforms and somewhat lower for RNA-Seq, yet 95% confidence intervals of AUC statistics overlapped among platforms.
The correlation of fold changes derived from high-throughput platforms and qRT-PCR (reference method) were higher for sequencing methods than microarrays and especially for DriverMap (Fig. 3). The estimated R2 coefficient for correlation of Salmon40 quantified RNA-Seq expression and qRT-PCR expression changes in a two group analysis (TIL vs TNL) was identical to the one previously reported (0.85) based on wet-lab validation of 18,080 protein-coding genes in human brain tissues24 and relying on RNA-Seq data generated in the FDA sponsored Sequencing Quality Control (SEQC) study12,16. The more compressed fold-changes derived from microarrays than with sequencing based techniques was expected due to the background levels owing to cross-hybridization37,38. Of interest, for changes associated with labor at term, the dynamic range of DriverMap exceeded the one of qRT-PCR, with the slope in the log2 fold change correlation plot significantly exceeding parity (slope of 1.13, which is significantly >1.0).
Comparisons to other studies of transcriptomic changes with gestation
The impact of advancing gestation on the maternal whole blood transcriptome in normal pregnancies has been previously evaluated, and results vary depending on the intervals of gestation considered, sample size, expression profiling platforms, and significance cut-offs. Heng et al.21 reported only 45 genes changing with gestation from 17–23 to 27–33 weeks. This number increases to 2,321 if the cut-off on the magnitude of change is removed and hence genes are selected based only on adjusted p-values. Al-Garawi et al.22 reported that 12% of the transcriptome (3,830 unique genes) is modulated during gestation, yet in this later study, the span of gestation was much larger, including first-trimester and term gestation samples (10–18 weeks vs 30–38 weeks). This larger fraction of genes changing with gestation is rather similar to the one of maternal plasma proteins that we have previously reported to change with gestational age, using frequent sampling from early to term gestation41. All three transcriptomics platforms evaluated in this study identified significantly more genes than expected by chance among those reported previously. Although enrichment ORs were higher for the HTA microarrays, DriverMap confirmed more of the previously reported gene changes and provided the strongest evidence for an enrichment (smallest p-values) (Table 1).
The importance of identifying transcripts associated with gestational age was recently highlighted in a study by Ngo et al.8 who screened cell free RNA transcripts among those that are placental, immune, and fetal-liver specific, with the goal of developing a prediction model for the interval from blood draw to term delivery. Of the 20 immune and placenta-specific genes highlighted as changing with gestation by Ngo et al.8 and also detected on all three platforms in maternal whole blood, DriverMap identified five (ANXA3, ARG1, S100A8, S100P, and ADAM12) as being significantly modulated during gestation, followed by HTA microarrays and RNA-Seq that identified three and one of these genes, respectively.
Cell type-specific gene expression signatures as markers for placental function
Gene set analysis is one of the few means available to researchers to interpret omics studies and translate omics findings across platforms and species14,42,43. When the number of differentially expressed genes identified in a given condition is large, gene set analysis can be used to identify a few categories of genes that share a similar function and that are over-represented/enriched within the list of differentially expressed genes44. By contrast, when no significant changes can be demonstrated at the gene level, for example, due to a modest effect or small sample size, gene set analysis can use modest but coordinated changes in expression to establish a link between the phenotype and a predefined group of functionally related genes45. Another approach that implements gene set information to increase statistical power is the use of a gene set level summary as a biomarker46. In pregnancy research, Tsang et al.2 characterized the expression patterns of subpopulations of placental cells, including some of fetal (e.g., extravillous trophoblast and syncytiotrophoblast), maternal (e.g., decidual cells) and mixed origin (e.g., T cells). The authors have defined cell type-specific genes as those having higher expression in a given cell type compared to all others in the population of cells, similar in concept to defining tissue-specific gene sets47,48. The cell-free RNA expression (normalized sequence count) average over genes in the extravillous trophoblast signature was shown to be elevated in plasma of women diagnosed with preeclampsia compared to those with normal pregnancy. The authors have also shown that, of all cell-type signatures considered, the expression of B-cell and T-cell signatures changed most markedly with gestational age. While B-cell signature decreased monotonically from the first to the third trimester, T-cell signature decreases from the first to the second trimester and then increased during the third trimester. Despite differences between these two studies in terms of the type of samples (plasma vs whole blood) and downstream methods for RNA preservation and separation/extraction, our data strongly support these expression patterns with gestational age. A possible explanation for these similarities is that the plasma cell-free RNA included transcripts released by the white blood cells (cellular transcriptome reported herein). We also demonstrated that targeted expression profiling by DriverMap is especially suitable to track cell type-specific signatures as it leads to stronger associations and expression summaries that correlate better with qRT-PCR results (Figs 4 and 5).
T-cell signature as a marker for onset of labor
This work demonstrated for the first time a significant increase in the T-cell expression signature in maternal whole blood from women who underwent spontaneous labor at term compared to those who delivered at term without labor. This observation is in line with evidence supporting a role for maternal T cells in the physiologic and pathologic processes of term and preterm labor, including: (1) effector and activated T cells are found at the maternal-fetal interface before49–55 and during the physiologic process of labor56–58; (2) effector T cells are present in the peripheral blood before59–61 and during labor62; (3) the absence of T cells results in increased susceptibility to endotoxin-induced preterm labor, which was reversed by the adoptive transfer of CD4+ T cells63; (4) histopathological lesions characterized by the infiltration of maternal T cells into the placental tissues (i.e., villitis of unknown etiology64–66, chronic chorioamnionitis67, and chronic deciduitis68) are associated with preterm labor and labor at term67,69–73; (5) effector and regulatory T-cell subsets at the maternal-fetal interface are associated with the timing of term parturition74 and the onset of preterm labor75–77; and (6) in vivo T-cell activation causes the onset of preterm labor78. More recently, we reported that fetal T cells can also participate in the mechanisms that lead to spontaneous preterm labor by responding toward maternal antigens and releasing pro-inflammatory cytokines79. Collectively, these data indicate that T cells are implicated in the physiological and pathological processes of labor and that sequencing-based techniques that quantify cell type-specific signatures in maternal whole blood could provide a read-out of the immunological events taking place at the maternal-fetal interface and in the placenta.
Conclusions
Sequencing-based techniques are more suitable to quantify whole blood gene expression compared to microarrays, as they have an expanded dynamic range and identify more true positives. Targeted expression profiling by DriverMap identified more differentially expressed genes with gestational age and with labor at term than the other two platforms, and it is more accurate than untargeted RNA-Seq in measuring protein-coding gene expression, as it achieves higher coverage for coding genes for a lower total number of sequenced reads per sample. We have also demonstrated that targeted expression profiling is especially suited to track cell type-specific signatures discovered by single-cell transcriptomics in placental tissues and showed that maternal whole blood could provide a readout of the immunological events taking place at the maternal-fetal interface and in the placenta.
Methods
Study design
The study was part of a prospective longitudinal study that enrolled women with a normal pregnancy attending the Center for Advanced Obstetrical Care and Research of the Perinatology Research Branch, NICHD/NIH/DHHS, and the Detroit Medical Center/Wayne State University from February 2011 to May 2015. For gene expression profiling, we selected normal pregnancies with (n = 8) and without (n = 8) spontaneous labor at term. For one-half of the women in each labor group, we profiled three longitudinal samples collected at 12 to 40 weeks of gestation (Fig. 4), while for the other one-half of the women in each labor group, we profiled one sample taken at term before delivery (Fig. 5), for a total of 32 transcriptomes (Table S1). All patients provided written informed consent. The use of biological specimens as well as clinical data for research purposes was approved by the Institutional Review Boards of Wayne State University and NICHD. All experiments were performed in accordance with relevant guidelines and regulations.
Sample collection and processing
Whole blood samples were collected directly into PAXgene Blood RNA tubes, stored at room temperature for 24 hours, and frozen at −80 °C until analysis. The PAXgene tubes contain a stabilizing additive to maintain cellular RNA integrity for extended periods of time; cellular RNA integrity is reported to be preserved up to 5 years80.
RNA Extraction
RNA was isolated in April 2016 from PAXgene® Blood RNA collection tubes (BD 762165), as described in the PAXgene® Blood miRNA Kit Handbook (December, 2015). Purified RNA was quantified by UV spectrophotometry using the DropSense96® Microplate Spectrophotometer (Trinean) and quality assessed by microfluidics using the RNA ScreenTape on the Agilent 2200 TapeStation.
Microarray profiling
A quantity of 100 ng of RNA was reverse-transcribed and amplified using the Affymetrix WT Plus Expression Kit (needs vendor information), following the manufacturer’s suggested protocol. A quantity of 5.5 μg of sense strand cDNA was fragmented and labeled using the Affymetrix WT Terminal Labeling Kit. 200 μl of labeled targets were hybridized to Affymetrix Human Transcriptome Arrays 2.0 GeneChip in an Affymetrix hybridization oven at 45 °C at 60 rpm for 16 hours. Washing and staining were performed on an Affymetrix Fluidics Station 450 and scanned on an Affymetrix GeneChip scanner 3000. Raw intensity data were generated from array images using Affymetrix AGCC software.
RNA-Seq profiling
Starting with 500 ng of total RNA, cDNA library templates were synthesized using the Illumina TruSeq® Stranded Total RNA LT (Set A) Kit with Ribo-Zero Globin rRNA reduction, as described in the TruSeq® Stranded Total RNA Sample Preparation Guide (Rev. E., October 2013). Libraries were validated using the HS D1000 ScreenTape on the Agilent 2200 TapeStation and quantified on the Qubit® 2.0 Fluorometer by the Qubit® dsDNA HS Assay (ThermoFisher). The Illumina HiSeq2500 instrument was used to cluster the samples onto the flow cell. The pools were put on at 10 pM and run in rapid mode at 2 × 100 paired end. The Illumina HiSeq Rapid PE Cluster Kit V2 and HiSeq Rapid SBS Kit v2 200 cycles were used.
Targeted expression profiling by DriverMap
The DriverMap Human Genome-Wide Gene Expression Profiling Assay (hDM18Kv2; Cellecta Inc., Mountain View, CA) was used to measure the expression level of 18,559 protein coding genes by combining highly multiplexed RT-PCR amplification with Next-Generation Sequencing quantitation. Sample processing was performed per manufacturer protocols available at https://www.cellecta.com/technology-portfolio/targeted-expression-profiling-driver-map-assay/. cDNA products amplified in the assay were analyzed on an Illumina NextSeq. 500 sequencer using a NextSeq500/550 High Output v2 Kit (75 cycles).
qRT-PCR profiling
Total RNA (150 ng/sample) was reverse-transcribed into cDNA using Reverse Transcription Master Mix (100–6298, Fluidigm, San Francisco, CA). The reaction system included 150 ng of sample total RNA and 1 μl of the master mix plus RNase-free water to bring the final volume to 5 μl. The BioMark System (Fluidigm) was used to perform high-throughput qPCR. For this system, specific target amplification of cDNA was performed. Briefly, a 0.2X pool of specific TaqMan gene expression assays (Applied Biosystems) was prepared by mixing the individual 20X assays (total 93, 1 μl of each) and 7 μl of RNase-free water and used as the source of primers. Pre-amplification reactions contained 1.25 μl of cDNA, 2.5 μl of TaqMan PreAmp Master Mix (4391128, Applied Biosystems), and 1.25 μl of the pooled TaqMan assay mix. The reaction was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems) for 14 cycles at 95 °C for 15 seconds and at 60 °C for 4 minutes. After the cycling, the pre-amplicons were diluted 1:5 with RNase-free water to a final volume of 25 μl. A Fluidigm 96.96 Dynamic Array chip was used to perform the qPCR assays. The chip was primed in an integrated fluidic circuit controller. After the priming, 2.75 μl of 20X TaqMan gene expression assay (Applied Biosystems) were mixed with 2.75 μl of 2X assay loading reagent (100–7611, Fluidigm) individually and loaded into the assay inlet on the chip; 2.25 μl of preamplified cDNA were mixed with 2.5 μl of TaqMan Universal PCR Master Mix (4304437, Applied Biosystems) and 0.25 μl of 20X GE sample loading reagent (100–7610, Fluidigm) and loaded into the sample inlet on the chip. The chip was returned to the integrated fluidic circuit controller for loading. After the samples and assays were loaded, the chip was placed into the BioMark System to amplify the target genes. The cycle threshold (Ct) value of each reaction on the chip was obtained with the Fluidigm RT-PCR analysis software.
Data analysis
Microarray data preprocessing
Human Transcriptome Arrays contain >6.0 million distinct probes grouped into probesets that target the exonic regions of 245,349 coding and 40,914 non-coding transcripts drawn from multiple data sources (RefSeq, Ensembl, UCSC, etc.). Microarray raw gene expression data were background corrected, quantile normalized, and summarized for each of the 44,699 coding and 22,829 corresponding transcript clusters (genes) using Robust Multi-array Average (RMA)81 implemented in the oligo package82 using probe to transcript cluster assignments from the hta20sttranscriptcluster.db package of Bioconductor83. P-values for expression above background levels for each probeset targeting individual exons of the genes were obtained using the Affymetrix Expression ConsoleTM version 1.4. Identification of genes as protein coding was based on HTA-2_0.na36.hg19.transcript annotation provided by the manufacturer. Microarray data were deposited in the Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113809.
Preprocessing RNA-Seq data
Paired-end RNA-Seq sequence data in fastq format were processed using Salmon aligner (version 0.9.1)40 in quasi-mapping-based mode using the Ensembl GRCh37.75 (hg19) version of the transcriptome that included both coding and non-coding genes. Expression quantification included correction for sequence-specific biases and fragment-level GC biases to generate counts per million for each transcript scaled up to library size. Sequence count data inferred from Salmon gene abundance were imported into R using the tximport package84. Raw and processed data from RNA-Seq experiments are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113964.
Pre-processing targeted RNA-Seq data
Representation of gene-specific amplicons in amplified cDNA products was analyzed using custom gene enumeration software developed by Cellecta, Inc. With this method, only reads that align to the gene-specific amplicon are counted. The alignment uses only the 36 nucleotides started by forward primer position and 36 nucleotides region ended by reverse primer. Any reads from the middle portion of the amplicons are not counted. To determine the impact of the expression quantification procedure, the exact same procedure based on Salmon aligner40 was also used to generate alternative count summaries for DriverMap. Raw and processed data for DriverMap sequencing experiments are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114037.
Quantification of cell type-specific expression
Thirteen cell type-specific gene sets (3 to 27 genes each) identified in placental single-cell genomics2 were summarized in each sample by averaging expression data (log2 normalized intensity for microarrays, log2 normalized counts for RNA-Seq and DriverMap, and −ΔCt for qRT-PCR).
Differential expression analysis
Gene expression data from all four platforms (microarray, RNA-Seq, targeted expression profiling, and qRT-PCR) were analyzed to quantify the effect of gestational age and the effect of labor at term on gene expression.
The effect of gestation was assessed within subject by performing a paired analysis contrasting expression data in one sample at term (gestational age ≥37 weeks) against the two samples collected preterm (<37 weeks). This analysis was restricted to the eight women who had three longitudinal measurements. To quantify changes with gestational age, we used linear models in which the response variable was the expression of each gene, while independent variables included the gestational age as a binary variable (term vs preterm gestation) as well as a subject-specific fixed effect, hence implementing a paired (within subject) analysis.
The effect of spontaneous labor at term was evaluated by performing an unpaired analysis of expression data in samples collected at term between eight women with term labor (TIL) and eight women who delivered at term without spontaneous labor (TNL). Linear models used to assess the effect of spontaneous labor at term included the expression of each gene as response variable and the group (TIL vs TNL) as an independent variable.
Microarray gene expression data were analyzed using linear models implemented in the limma package85 of Bioconductor. qRT-PCR data were analyzed using the same type of models starting with −ΔCt values, which are surrogates of log2 expression normalized with respect to housekeeping genes (ACTB, B2M, GAPDH, POLR2A, RPL37A, and RPLPO).
Count data obtained for the two sequencing based platforms (RNA-Seq and DriverMap) were analyzed using negative binomial models implemented in the DESeq. 2 package35,86 according to the same independent variables described for microarray and qRT-PCR data.
For all three high-throughput gene expression platforms, differences were considered significant if the false discovery rate-adjusted p-values (q-value) were <0.1, given that the transcripts were deemed expressed. The less conservative false discovery rate cut-off (10% as opposed to 5%) was selected to provide increased power to assess differences in the number of genes differentially expressed with a given phenotype by the three expression profiling methods, and also to quantify the overlap with lists of genes reported in other studies. For microarrays, transcript clusters were considered expressed if at least one probeset targeting the same transcript cluster had a detection p-value < 0.05 in at least 5 of the 32 samples. For sequencing based methods, a raw sequence count ≥5 in at least 5 of the 32 samples was required to call a gene expressed.
Selection of candidate genes for validation
The design of the qRT-PCR validation study was based on a preliminary analysis of the high-throughput data and aimed to include the top 10 genes identified by only one method and all genes identified by all three methods in each of the two comparisons (effect of gestational age and effect of labor). The preliminary analysis differed from the one reported above in terms of 1) criteria used to define expression above background for microarrays (using a cut-off for absolute intensity as opposed to detection p-values), 2) did not involve the exclusion of the contaminated sample for DriverMap analysis, and 3) used different differential expression criteria [nominal p < 0.005 and absolute log2 ratio > log2 (1.5)]. The assay identifiers for the target and reference genes used in the qRT-PCR experiments are shown in Table S3.
Ethics approval and consent to participate
All patients provided written informed consent. The use of biological specimens as well as clinical data for research purposes were approved by the Institutional Review Boards of Wayne State University and NICHD.
Electronic supplementary material
Acknowledgements
We thank Dr. Zhong Dong for assistance selecting the qRT-PCR primers for validation and running the validation experiments. We thank Dr. Zhou Ming (NIH/NCI) for overseeing the HTA microarray analysis at the National Cancer Institute at Frederick, Maryland. We thank Dan Lot for running the RNA-Seq experiments at the Applied Genomics Technology Center of Wayne State University in Detroit, Michigan. We acknowledge Alex Chenchik from Cellecta Inc., Mountain View, California, for providing the DriverMap assay service at no cost to the investigators. This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS); and, in part, with Federal funds from NICHD/NIH/DHHS under Contract No. HHSN275201300006C. ALT was also supported by the Perinatal Initiative of the Wayne State University School of Medicine. Dr. Romero has contributed to this work as part of his official duties as an employee of the United States Federal Government.
Author Contributions
A.L.T., R.R. and V.J.C. conceived the research. A.L.T. and Z.X. carried out research. A.L.T. drafted the manuscript. V.J.C. provided feedback on data analysis. A.L.T., Z.X., N.G.L., S.S.H., R.R., O.E., C.D.H. and V.J.C. reviewed and edited the manuscript. All authors read and approved the final manuscript.
Data Availability
The raw and summarized expression data for microarrays, RNA-Seq and targeted expression profiling by RNA-Seq are available as a Gene Expression Omnibus super series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113966). The R analysis script and data required to reproduce the main figures and tables is available from the authors’ website at http://bioinformaticsprb.med.wayne.edu/software/.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Adi L. Tarca, Email: atarca@med.wayne.edu
Roberto Romero, Email: prbchiefstaff@med.wayne.edu.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36649-w.
References
- 1.Hong X, et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:2467–2472. doi: 10.1073/pnas.1719264115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsang JCH, et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E7786–E7795. doi: 10.1073/pnas.1710470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin JZ, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nature methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souza MF, et al. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PloS one. 2017;12:e0184094. doi: 10.1371/journal.pone.0184094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngo TTM, et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science. 2018;360:1133–1136. doi: 10.1126/science.aar3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic acids research. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosens I, Pijnenborg R, Vercruysse L, Romero R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. American journal of obstetrics and gynecology. 2011;204:193–201. doi: 10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Walter E, Stenger D, Thach D. Effects of globin mRNA reduction methods on gene expression profiles from whole blood. The Journal of molecular diagnostics: JMD. 2006;8:551–558. doi: 10.2353/jmoldx.2006.060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium M, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nature biotechnology. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarca AL, et al. Strengths and limitations of microarray-based phenotype prediction: lessons learned from the IMPROVER Diagnostic Signature Challenge. Bioinformatics. 2013;29:2892–2899. doi: 10.1093/bioinformatics/btt492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dayarian A, et al. Predicting protein phosphorylation from gene expression: top methods from the IMPROVER Species Translation Challenge. Bioinformatics. 2015;31:462–470. doi: 10.1093/bioinformatics/btu490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarac OS, et al. Species translatable blood gene signature as a marker of exposure to smoking: computational approaches of the top ranked teams in the sbv IMPROVER Systems Toxicology challenge. Comput Toxicol. 2018;5:25–30. doi: 10.1016/j.comtox.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium SM-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nature biotechnology. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng M, et al. A benchmark for RNA-seq quantification pipelines. Genome biology. 2016;17:74. doi: 10.1186/s13059-016-0940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peixoto L, et al. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic acids research. 2015;43:7664–7674. doi: 10.1093/nar/gkv736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: Visualization of Intersecting Sets. IEEE transactions on visualization and computer graphics. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]
- 21.Heng YJ, et al. Maternal Whole Blood Gene Expression at 18 and 28 Weeks of Gestation Associated with Spontaneous Preterm Birth in Asymptomatic Women. PloS one. 2016;11:e0155191. doi: 10.1371/journal.pone.0155191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Garawi A, et al. The Role of Vitamin D in the Transcriptional Program of Human Pregnancy. PloS one. 2016;11:e0163832. doi: 10.1371/journal.pone.0163832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwemer LM, Hui L, Wick HC, Bianchi DW. RNA-Seq and expression microarray highlight different aspects of the fetal amniotic fluid transcriptome. Prenatal diagnosis. 2014;34:1006–1014. doi: 10.1002/pd.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everaert C, et al. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Scientific reports. 2017;7:1559. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archer KJ, Reese SE. Detection call algorithms for high-throughput gene expression microarray data. Briefings in bioinformatics. 2010;11:244–252. doi: 10.1093/bib/bbp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaiworapongsa T, et al. Differences and similarities in the transcriptional profile of peripheral whole blood in early and late-onset preeclampsia: insights into the molecular basis of the phenotype of preeclampsiaa. Journal of perinatal medicine. 2013;41:485–504. doi: 10.1515/jpm-2013-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paquette AG, et al. Comparative analysis of gene expression in maternal peripheral blood and monocytes during spontaneous preterm labor. American journal of obstetrics and gynecology. 2018;218:345 e341–345 e330. doi: 10.1016/j.ajog.2017.12.234. [DOI] [PubMed] [Google Scholar]
- 29.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nature reviews. Genetics. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 30.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pineles BL, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. American journal of obstetrics and gynecology. 2007;196:261 e261–266. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Romero R, et al. Transcriptome interrogation of human myometrium identifies differentially expressed sense-antisense pairs of protein-coding and long non-coding RNA genes in spontaneous labor at term. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2014;27:1397–1408. doi: 10.3109/14767058.2013.860963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montenegro D, et al. Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition. The Journal of pathology. 2009;217:113–121. doi: 10.1002/path.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gormley M, et al. Preeclampsia: novel insights from global RNA profiling of trophoblast subpopulations. American journal of obstetrics and gynecology. 2017;217:200 e201–200 e217. doi: 10.1016/j.ajog.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Royce TE, Rozowsky JS, Gerstein MB. Toward a universal microarray: prediction of gene expression through nearest-neighbor probe sequence identification. Nucleic acids research. 2007;35:e99. doi: 10.1093/nar/gkm549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okoniewski MJ, Miller CJ. Hybridization interactions between probesets in short oligo microarrays lead to spurious correlations. BMC bioinformatics. 2006;7:276. doi: 10.1186/1471-2105-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nature methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero R, et al. The maternal plasma proteome changes as a function of gestational age in normal pregnancy: a longitudinal study. American journal of obstetrics and gynecology. 2017;217:67 e61–67 e21. doi: 10.1016/j.ajog.2017.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarca AL, Bhatti G, Romero R. A comparison of gene set analysis methods in terms of sensitivity, prioritization and specificity. PloS one. 2013;8:e79217. doi: 10.1371/journal.pone.0079217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nature genetics. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 45.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 46.Ozerov IV, et al. In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nature communications. 2016;7:13427. doi: 10.1038/ncomms13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarui T, et al. Amniotic fluid transcriptomics reflects novel disease mechanisms in fetuses with myelomeningocele. American journal of obstetrics and gynecology. 2017;217:587 e581–587 e510. doi: 10.1016/j.ajog.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sindram-Trujillo A, Scherjon S, Kanhai H, Roelen D, Claas F. Increased T-cell activation in decidua parietalis compared to decidua basalis in uncomplicated human term pregnancy. American journal of reproductive immunology. 2003;49:261–268. doi: 10.1034/j.1600-0897.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 50.Sindram-Trujillo AP, et al. Comparison of decidual leukocytes following spontaneous vaginal delivery and elective cesarean section in uncomplicated human term pregnancy. Journal of reproductive immunology. 2004;62:125–137. doi: 10.1016/j.jri.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Tilburgs T, et al. Differential distribution of CD4(+)CD25(bright) and CD8(+)CD28(−) T-cells in decidua and maternal blood during human pregnancy. Placenta. 2006;27(Suppl A):S47–53. doi: 10.1016/j.placenta.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Tilburgs T, Scherjon SA, Roelen DL, Claas FH. Decidual CD8+ CD28− T cells express CD103 but not perforin. Human immunology. 2009;70:96–100. doi: 10.1016/j.humimm.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Tilburgs T, et al. Expression of NK cell receptors on decidual T cells in human pregnancy. Journal of reproductive immunology. 2009;80:22–32. doi: 10.1016/j.jri.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Tilburgs T, et al. Human decidual tissue contains differentiated CD8+ effector-memory T cells with unique properties. Journal of immunology. 2010;185:4470–4477. doi: 10.4049/jimmunol.0903597. [DOI] [PubMed] [Google Scholar]
- 55.Powell RM, et al. Decidual T Cells Exhibit a Highly Differentiated Phenotype and Demonstrate Potential Fetal Specificity and a Strong Transcriptional Response to IFN. Journal of immunology. 2017;199:3406–3417. doi: 10.4049/jimmunol.1700114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez-Lopez N, et al. Specific inflammatory microenvironments in the zones of the fetal membranes at term delivery. Am J Obstet Gynecol. 2011;205(235):e215–224. doi: 10.1016/j.ajog.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 57.Gomez-Lopez N, Hernandez-Santiago S, Lobb AP, Olson DM, Vadillo-Ortega F. Normal and premature rupture of fetal membranes at term delivery differ in regional chemotactic activity and related chemokine/cytokine production. Reproductive sciences. 2013;20:276–284. doi: 10.1177/1933719112452473. [DOI] [PubMed] [Google Scholar]
- 58.Gomez-Lopez N, et al. Evidence for a role for the adaptive immune response in human term parturition. American journal of reproductive immunology. 2013;69:212–230. doi: 10.1111/aji.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Y, et al. Peripheral CD300a+ CD8+ T lymphocytes with a distinct cytotoxic molecular signature increase in pregnant women with chronic chorioamnionitis. American journal of reproductive immunology. 2012;67:184–197. doi: 10.1111/j.1600-0897.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aghaeepour, N. et al. An immune clock of human pregnancy. Science immunology2, 10.1126/sciimmunol.aan2946 (2017). [DOI] [PMC free article] [PubMed]
- 61.Shah NM, et al. Changes in T Cell and Dendritic Cell Phenotype from Mid to Late Pregnancy Are Indicative of a Shift from Immune Tolerance to Immune Activation. Frontiers in immunology. 2017;8:1138. doi: 10.3389/fimmu.2017.01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan M, Jordan F, McInnes IB, Harnett MM, Norman JE. Leukocytes are primed in peripheral blood for activation during term and preterm labour. Molecular human reproduction. 2009;15:713–724. doi: 10.1093/molehr/gap054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bizargity P, Del Rio R, Phillippe M, Teuscher C, Bonney EA. Resistance to lipopolysaccharide-induced preterm delivery mediated by regulatory T cell function in mice. Biology of reproduction. 2009;80:874–881. doi: 10.1095/biolreprod.108.074294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JS, et al. Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology. Histopathology. 2008;52:457–464. doi: 10.1111/j.1365-2559.2008.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim MJ, et al. Villitis of unknown etiology is associated with a distinct pattern of chemokine up-regulation in the feto-maternal and placental compartments: implications for conjoint maternal allograft rejection and maternal anti-fetal graft-versus-host disease. Journal of immunology. 2009;182:3919–3927. doi: 10.4049/jimmunol.0803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito Y, et al. Increased expression of perforin, granzyme B, and C5b-9 in villitis of unknown etiology. Placenta. 2015;36:531–537. doi: 10.1016/j.placenta.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 67.Kim CJ, et al. The frequency, clinical significance, and pathological features of chronic chorioamnionitis: a lesion associated with spontaneous preterm birth. Mod Pathol. 2010;23:1000–1011. doi: 10.1038/modpathol.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khong TY, et al. Chronic deciduitis in the placental basal plate: definition and interobserver reliability. Human pathology. 2000;31:292–295. doi: 10.1016/S0046-8177(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 69.Lee J, et al. A signature of maternal anti-fetal rejection in spontaneous preterm birth: chronic chorioamnionitis, anti-human leukocyte antigen antibodies, and C4d. PLoS One. 2011;6:e16806. doi: 10.1371/journal.pone.0016806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J, et al. Chronic chorioamnionitis is the most common placental lesion in late preterm birth. Placenta. 2013;34:681–689. doi: 10.1016/j.placenta.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 71.Kim CJ, Romero R, Chaemsaithong P, Kim JS. Chronic inflammation of the placenta: definition, classification, pathogenesis, and clinical significance. Am J Obstet Gynecol. 2015;213:S53–69. doi: 10.1016/j.ajog.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maymon E, et al. Chronic inflammatory lesions of the placenta are associated with an up-regulation of amniotic fluid CXCR3: A marker of allograft rejection. J Perinat Med. 2018;46:123–137. doi: 10.1515/jpm-2017-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamblyn JA, Lissauer DM, Powell R, Cox P, Kilby MD. The immunological basis of villitis of unknown etiology - review. Placenta. 2013;34:846–855. doi: 10.1016/j.placenta.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 74.Gomez-Lopez N, Olson DM, Robertson SA. Interleukin-6 controls uterine Th9 cells and CD8(+) T regulatory cells to accelerate parturition in mice. Immunol Cell Biol. 2016;94:79–89. doi: 10.1038/icb.2015.63. [DOI] [PubMed] [Google Scholar]
- 75.Arenas-Hernandez M, et al. An imbalance between innate and adaptive immune cells at the maternal-fetal interface occurs prior to endotoxin-induced preterm birth. Cell Mol Immunol. 2016;13:462–473. doi: 10.1038/cmi.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.St Louis D, et al. Invariant NKT Cell Activation Induces Late Preterm Birth That Is Attenuated by Rosiglitazone. Journal of immunology. 2016;196:1044–1059. doi: 10.4049/jimmunol.1501962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomez-Lopez N, et al. In vivo activation of invariant natural killer T cells induces systemic and local alterations in T-cell subsets prior to preterm birth. Clin Exp Immunol. 2017;189:211–225. doi: 10.1111/cei.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gomez-Lopez N, et al. In vivo T-cell activation by a monoclonal alphaCD3epsilon antibody induces preterm labor and birth. American journal of reproductive immunology. 2016;76:386–390. doi: 10.1111/aji.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frascoli, M. et al. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-gamma and TNF-alpha. Sci Transl Med10, 10.1126/scitranslmed.aan2263 (2018). [DOI] [PMC free article] [PubMed]
- 80.Kim JH, et al. Comparison of three different kits for extraction of high-quality RNA from frozen blood. SpringerPlus. 2014;3:76. doi: 10.1186/2193-1801-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 82.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. doi: 10.12688/f1000research.7563.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smyth, G. K. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor (ed. Gentleman, R. et al.) 397–420 (Springer, 2012).
- 86.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature protocols. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and summarized expression data for microarrays, RNA-Seq and targeted expression profiling by RNA-Seq are available as a Gene Expression Omnibus super series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113966). The R analysis script and data required to reproduce the main figures and tables is available from the authors’ website at http://bioinformaticsprb.med.wayne.edu/software/.