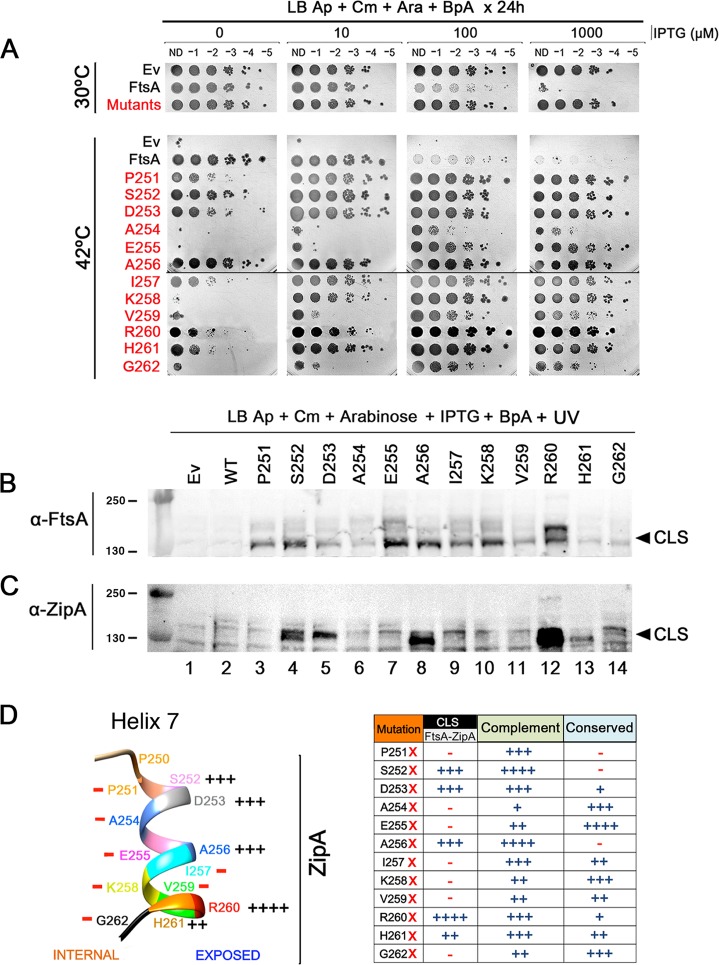
FIG 2.
Residues on the outer face of helix 7 of FtsA preferentially cross-link with ZipA. (A) WM1115 strains expressing FtsA amber substitutions at all the residues within helix 7 were grown in the presence of BpA and tested for their ability to complement ftsA12 at 42°C. After growth at 30°C, all strains shown were serially diluted 10-fold, spotted on plates containing different concentrations of IPTG to induce expression of the ftsA derivatives at different levels plus arabinose to induce pEVOL and BpA, and incubated either at 30°C or 42°C for 24 h. To save space, the “Mutants” row shows one of the mutants that grew at all IPTG levels at 30°C, because all of the other mutants behaved similarly. (B and C) The same strains in panel A were UV cross-linked, and protein extracts were separated by SDS-PAGE. Western blots were probed with either anti-FtsA (B) or anti-ZipA (C), and arrowheads indicate the general position of CLS. Molecular weight markers in kilodaltons are shown to the left. (D) Shown at the left is the position of each residue in FtsA helix 7 (including P250, which is outside the helix) and the relative ability of BpA at each position to cross-link with ZipA. The protein sequence of ZipA from E. coli was used to generate a model PDB file with the Phyre2 server and subsequently analyzed in UCSF Chimera. Summarized in the table at the right is the ability of each amber substitution under the conditions of the experiments in panels A and C to cross-link with ZipA and to complement at 42°C, along with the relative conservation of each substituted residue. The scoring system for CLS reflects the intensity of CLS bands, from ++++ (strongest) to − (undetectable). Scoring for complementation reflects the ability to grow at no IPTG like WT FtsA (++++), weak at no IPTG but strong growth with 10 µM IPTG (+++), weak with 10 µM IPTG but stronger growth at higher IPTG levels (++), or little growth at any induction level (+). Scoring for conservation depends on the degree of conservation across the species aligned in Fig. S5 in the supplemental material: ++++, identity across all species; +++, identity across most species; ++, identity in at least 4 other species in the list; +, some identity or similar characteristics in other species; −, no obvious conservation.