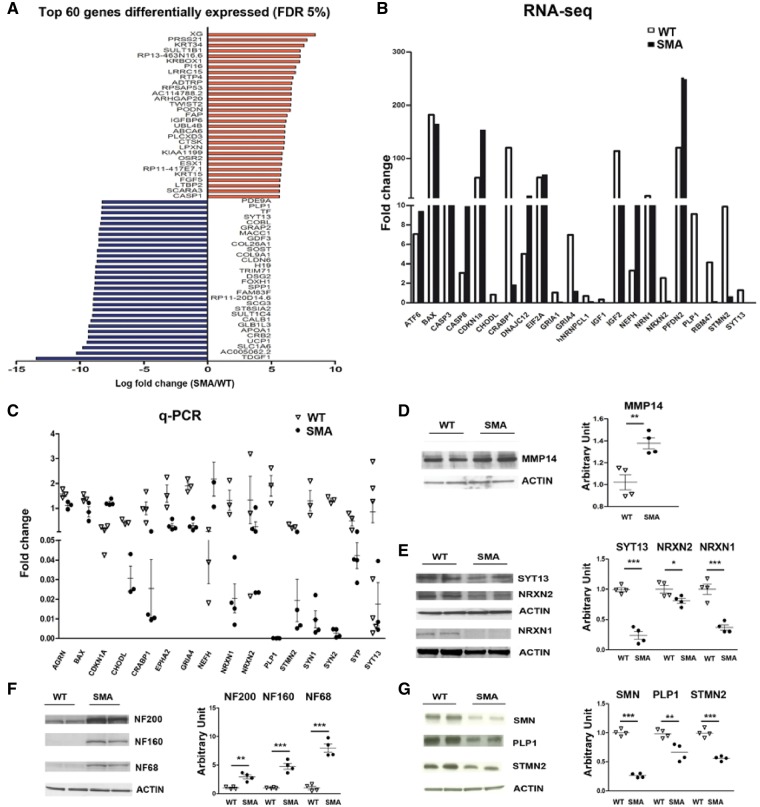
Figure 2.
Transcriptional analysis of SMA versus wild-type-motor neurons by RNA-Seq analysis shows deregulation of specific mRNAs involved in motor neuron functions. (A) Top 60 genes (based on fold change) differentially expressed in SMA versus wild-type motor neurons with an FDR of 5%. (B and C) Comparison of RNA-Seq and quantitative RT-PCR data or a selected number of transcripts in wild-type and SMA motor neurons. STMN2 (**P < 0.001), PLP1 (*P < 0.05), NEFH (*P < 0.05), NRXN1 (*P < 0.05), SYN1 (*P < 0.05), SYN2 (***P < 0.0001), EPH2A (*P < 0.05), SYP (*P < 0.05), CRABP1 (**P < 0.01), CDKN1A (***P < 0.0001), CHODL (*P < 0.05), GRIA4 (**P < 0.01). Student’s t-test. Error bars represent ± SEM from three independent experiments. (D–G) Representative image of western blot of selected proteins in SMA motor neurons compared to wild-type motor neurons: MMP14: **P < 0.01; SYT13: ***P < 0.0001; NRXN2: *P < 0.05; NRXN1: ***P < 0.001; NF200: **P < 0.01; NF160: ***P < 0.001; NF68: ***P < 0.001; SMN: ***P < 0.0001, PLP1: **P < 0.01; STMN2: ***P < 0.001, Student’s t-test. Data are presented as mean ± SEM from three independent experiments.