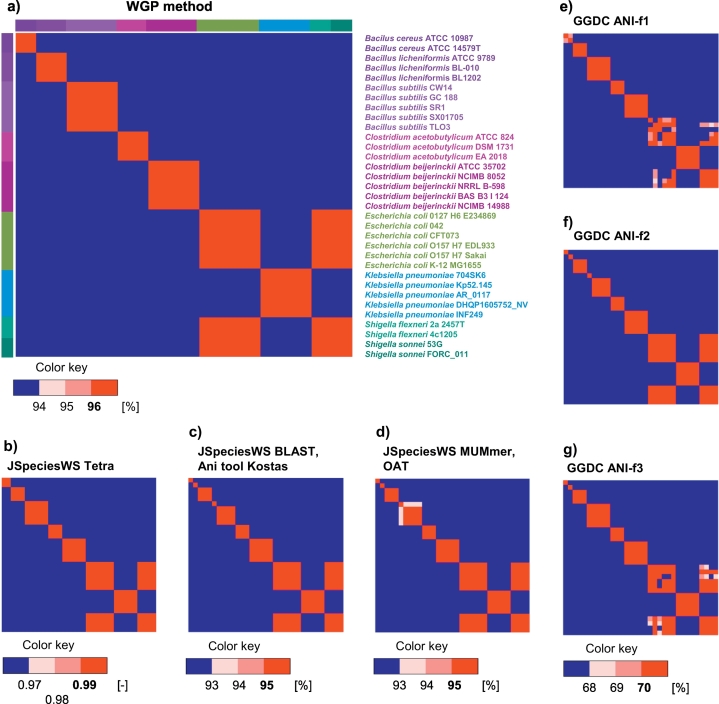
Fig. 6.
Visualization of the delineation results: a) our proposed WGP method; b) online tool JSpeciesWS using method TETRA; c) online tool JSpeciesWS using BLAST and Ani tool by Kostas lab; d) online tool JSpeciesWS using MUMmer and OAT; e) tool GGDC using formula ANI-f1; f) GGDC tool using the recommended formula ANI-f2; and g) GGDC tool using formula ANI-f3. The methods that have the same heatmap for the given threshold share one subplot. On the diagonal, there is a self-comparison of each genome where self-similarity is 100%. Similarity values above the threshold (bold value in the color key) are highlighted in red and similarity values 2% below the threshold are blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)