Figure 1.
sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing
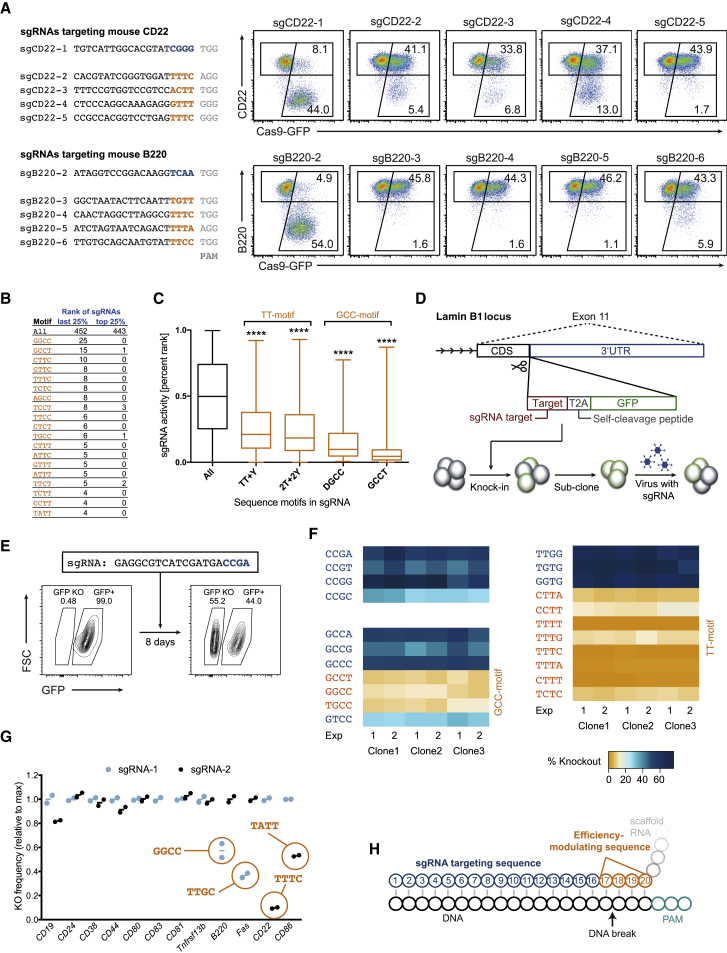
(A) Flow cytometry plots showing the knockout frequencies (lower right gate) of CD22 (top) and B220 (bottom) in primary B cells isolated from Cas9-transgenic (GFP+) animals 4 days post-transduction with retroviruses encoding the indicated sgRNAs. The sequences in the four PAM-proximal bases of the targeting sequence (hereafter called efficiency-modulating sequence [EMS]) are shown in orange (T-rich) or blue (control); the PAM is shown in gray. Data are representative for two independent experiments.
(B) Number of sgRNAs with the indicated sequences in the EMS in the inefficient (last 25%) or efficient (top 25%) sgRNAs reported by Doench et al. (2014).
(C) Boxplots of the sgRNA activities reported by Doench et al. (2014), considering all of the sgRNAs (black) or the sgRNAs with the indicated motifs in the EMS (orange). The top, middle, and bottom lines of the boxplot represent the 25th, 50th, and 75th percentiles, respectively; the whiskers represent the max and min values. Subgroups were compared to the control set using ordinary one-way ANOVA (∗∗∗∗p < 0.0001).
(D) Scheme of validation experiment. sgRNA target sequences followed by T2A and GFP were targeted in-frame into the Lamin B1 locus of a mouse B cell tumor cell line using CRISPR/Cas9. GFP+ cells were subcloned, transduced using retroviruses encoding the respective sgRNAs, and cultured for 8 days before analysis.
(E) Example of GFP knockout measurement (GFP KO gate) by flow cytometry 8 days post-transduction with the indicated sgRNA, as in the experimental system shown in (D).
(F) Heatmap of the GFP knockout frequencies in the cell lines with the indicated sequences in the EMS 8 days post-transduction with the respective sgRNAs. Sequences matching the TT- and GCC-motifs are shown in orange.
(G) Knockout frequencies in Cas9-transgenic primary B cells 4 days post-transduction with sgRNAs, normalized to the higher knockout frequency of the two sgRNAs used. The sgRNAs matching the TT- or GCC-motifs are encircled, and the respective sequences indicated. Data are based on two independent experiments and adapted from Chu et al. (2016).
(H) Schematic diagram of the targeting sequence of the sgRNA (blue and orange) bound to the DNA (black). The four PAM-proximal nucleotides of the targeting sequence (orange) were called EMS due to their potential for modulating knockout frequencies. The predicted CRISPR/Cas9-mediated DNA cut is indicated.