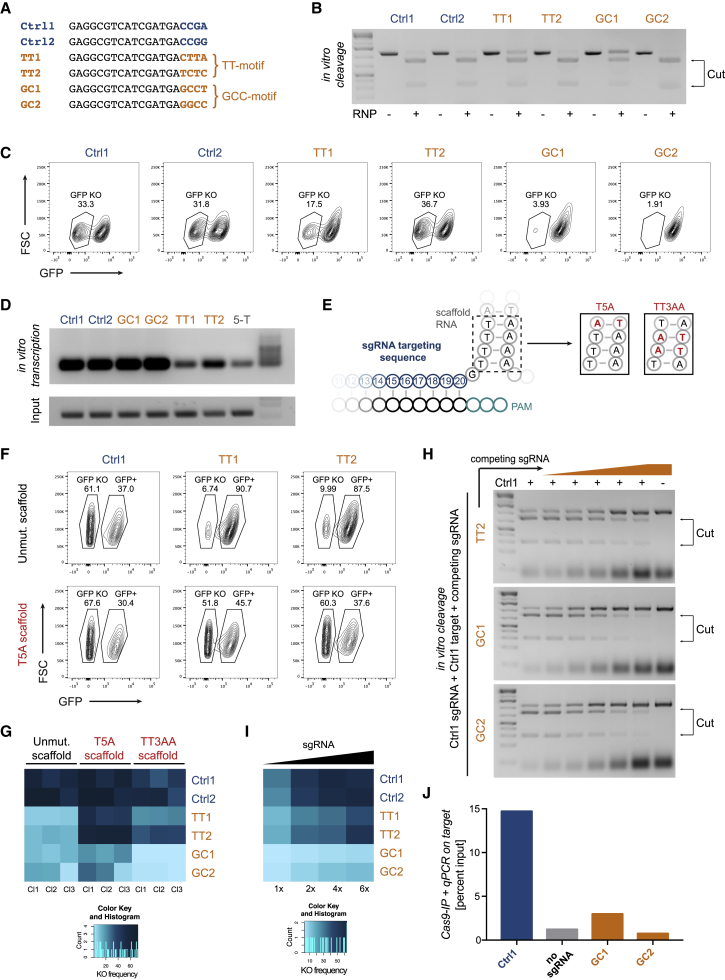
Figure 2.
Mechanism of Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing
(A) Targeting sequences of the sgRNAs used to study mechanism of motifs. The four PAM-proximal bases are highlighted in blue and orange.
(B) In vitro cleavage assay using ribonucleoprotein particles (RNPs) with the indicated sgRNAs and amplified target sequences.
(C) Knockout frequencies in vivo 2 days post-electroporation with the indicated synthetic sgRNAs.
(D) sgRNAs produced by in vitro transcription of the indicated sgRNAs and a negative control sgRNA having five Ts at the 3′ end of the targeting sequence (5-T).
(E) Scheme of the 3′ end of the targeting sequence and the 5′ end of the scaffold RNA. The four Ts in the scaffold were mutated to the indicated variants (T5A and TT3AA).
(F) Knockout frequencies 8 days post-transduction, with sgRNAs consisting of the indicated targeting sequences and variants of scaffold RNAs.
(G) Heatmap of the knockout frequencies obtained with the mutated scaffolds as in (F) in three clones (Cl) per condition.
(H) In vitro cleavage assay using Ctrl1 and the Ctrl1 target site in the presence of increasing levels (0×, 0.5×, 1×, 2×, 4×, 8×, and 8×) of the indicated competing sgRNAs.
(I) Heatmap of the knockout frequencies 8 days post-electroporation, with increasing doses of the indicated synthetic sgRNAs.
(J) Quantification of target sites bound to Cas9. Cas9 was immunoprecipitated 16 h post-transfection with sgRNA-encoding plasmids. Data are representative for two independent experiments.