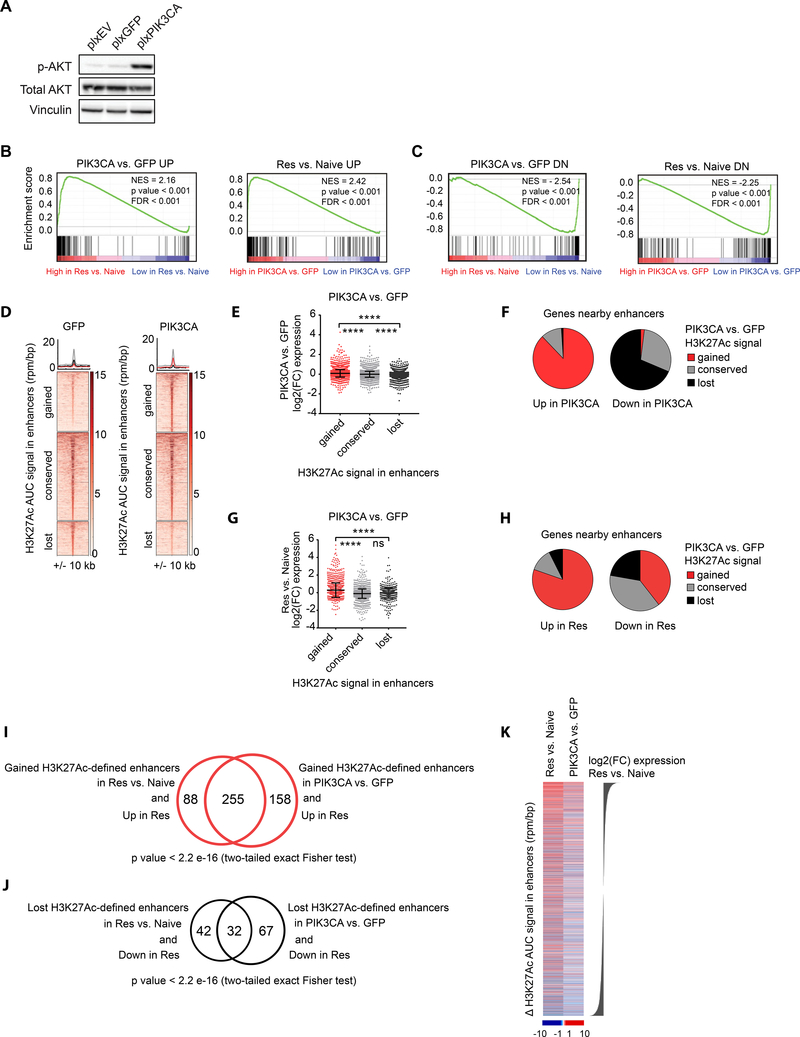
Figure 5: Transcriptomic analysis of BET inhibitor resistant cells reveals overexpression of PI3K signaling recapitulates enhancer remodeling and transcriptional changes characterizing the resistant state.
A. Western blot of SK-N-BE(2)-C cells engineered to overexpress an empty vector (plxEV), plxGFP or plxPIK3CA. B. GSEA demonstrating enrichment of genes upregulated in resistance among genes upregulated by PIK3CA overexpression (left) and vice versa (right). C. GSEA demonstrating enrichment of genes downregulated in resistance among genes downregulated by PIK3CA overexpression (left) and vice versa (right). D. Heatmaps showing H3K27Ac binding in gained, conserved and lost enhancer regions in PIK3CA vs. GFP samples. Each row represents a single genomic region +/− 10 kb from the enhancer center. Genomic occupancy is shaded by binding intensity in units of reads per million per base pair (rpm/bp). Regions are ranked by H3K27Ac binding signal in GFP cells. Metaplots for average binding intensities across the gained (red), conserved (gray) and lost (black) enhancer regions are shown on top. E. Dot plots showing log2(FC) in expression in PIK3CA vs. GFP cells for the genes associated with gained, conserved, and lost enhancers with PIK3CA overexpression (**** p value < 0.0001 un-paired two sample Student t-test with Welch correction). F. Pie charts showing the percentages of genes with gained, conserved or lost nearby enhancers with PIK3CA overexpression, among genes which are upregulated or downregulated by PIK3CA overexpression. G. Dot plots showing log2(FC) in expression in resistant vs. naive SK-N-BE(2)-C cells for the genes associated with gained, conserved, and lost enhancers with PIK3CA overexpression (**** p value < 0.0001, ns= not significant, un-paired two sample Student t-test with Welch correction). H. Pie charts showing the percentages of genes with gained, conserved and lost nearby enhancers with PIK3CA overexpression, among genes which are upregulated or downregulated in resistance. I. Venn-diagram showing the overlap of genes upregulated in resistant SK-N-BE(2)-C cells with nearby gained enhancers in resistance vs. genes upregulated in resistant SK-N-BE(2)-C cells nearby gained enhancers in PIK3CA overexpressing SK-N-BE(2)-C cells. Significance estimated based on two-tailed Fisher exact test. J. Venn-diagram showing the overlap of genes downregulated in resistant SK-N-BE(2)-C cells with nearby lost enhancers in resistant SK-N-BE(2)-C cells vs. genes downregulated in resistant SK-N-BE(2)-C cells with nearby lost enhancers in PIK3CA overexpressing SK-N-BE(2)-C cells. Significance estimated based on two-tailed Fisher exact test. K. Heatmaps showing ΔH3K27Ac AUC signal in enhancers for resistant vs. naive and PIK3CA vs. GFP samples ranked by log2(FC) expression in resistant vs. naive SK-N-BE(2)-C cells. Enhancers in this figure were defined by H3K37Ac binding. Dot plots in this figure are presented as mean values ± SD.