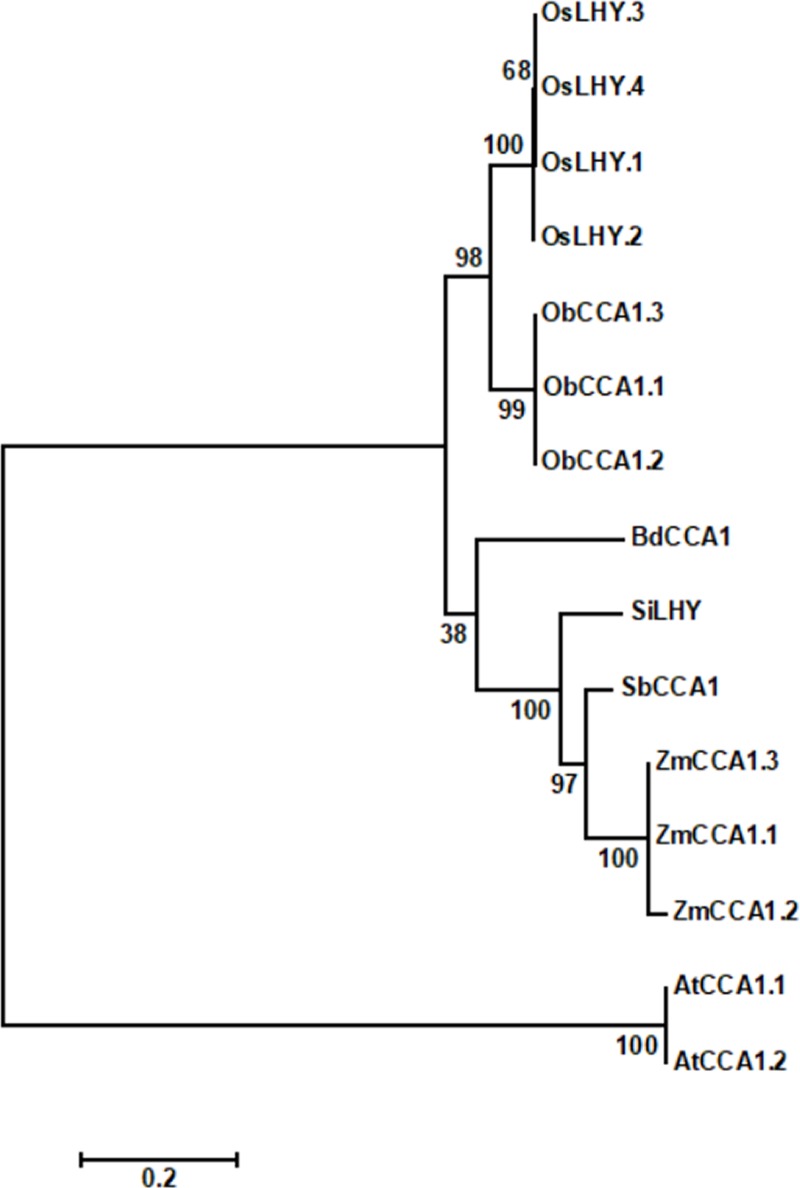
Fig 5. Phylogenetic analysis of CML288 ZmCCA1 splice variants and related genes.
Protein sequences were aligned by DNAMAN version 9.0. The phylogenetic tree was constructed using the maximum-likelihood method with full-length protein sequences. Species names and accession numbers are indicated: Zea mays, ZmCCA1 (ACG26686); A. thaliana, AtCCA1.1 (NP_850460), AtCCA1.2 (NP_001318437); Sorghum bicolor, SbCCA1 (LOC8080836); Setaria italica, SiLHY (LOC101764638); Oryza sativa Japonica Group, OsLHY.1 (XP_015649835), OsLHY.2 (XP_015649844), OsLHY.3 (XP_015649846), OsLHY.4 (XP_015649847); Oryza brachyantha, ObCCA1.1 (XP_006659138), ObCCA1.2 (XP_006659145), ObCCA1.3 (XP_015695906); Brachypodium distachyon, BdCCA1 (LOC100838310). Scale bar indicates 0.2 of amino acid substitutions per site.