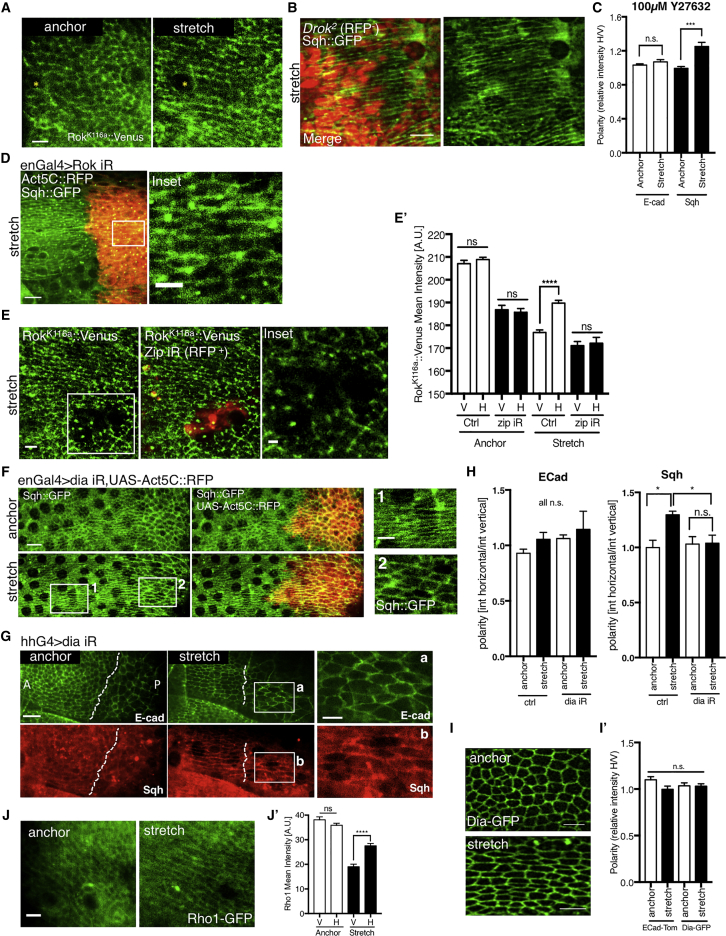
Figure 4.
Myosin II Polarity is Rok Independent and Depends on Dia Expression
(A) RokK116a::Venus localization in anchored and stretched third instar imaginal discs.
(B) Drok2 mitotic clones (RFP−) in Sqh::GFP third instar imaginal disc.
(C) Quantification of polarization for ECad::GFP and Sqh::Cherry expressing discs treated with Rok inhibitor Y27632 prior to stretching experiment. Statistical tests compare the change in polarity between the anchor and stretch states, n = 5.
(D) Sqh::GFP, enGal4>UAS-Rok-RNAi, and UAS-Act5CRFP discs subjected to stretching; inset demonstrates Sqh::GFP polarity in Rok-RNAi cells (marked by red).
(E) zip-RNAi clones (labeled as zip iR, indicated in red) in stretched third instar RokK116a::Venus discs. Quantifications of RokK116a::Venus mean intensity in (E’).
(F) Discs expressing Sqh::GFP and enGal4>UAS-dia-RNAi, UAS-Act5c-RFP that were stretched; UAS-Act5C::RFP marks dia iR (posterior side) in red. Insets compare Sqh::GFP polarity in stretched enGal4>dia iR and UAS-Act5c-RFP discs; panel 1 refers to anterior (ctrl) side; panel 2 refers to posterior (dia iR) side.
(G) Tissue marked with E-cad::GFP and Sqh::mCherry, expressing dia-RNAi (dia iR) with hhGal4 driver and subjected to stretching. Dotted line indicates A-P compartment boundary. Inset demonstrates lack of Sqh::GFP polarity in dia-RNAi cells.
(H) Quantification of E-cad::GFP polarity and Sqh::mCherry polarity in experiment described in (G). Polarity is measured as mean fluorescent intensity on horizontal junctions relative to vertical junctions (see STAR Methods for details); n = 4 wing discs.
(I) Dia::GFP localization in anchored and stretch third instar imaginal discs. (I′) Quantification of Dia::GFP polarity and ECad::tomato polarity in anchor (n = 3) and stretch (n = 6).
(J) Rho1::GFP protein trap localization in anchored and stretched third instar imaginal discs. (J′) Quantification of Rho1-GFP mean junctional intensity.
Error bars indicate S.E.M.; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 with t test. Scale bars, 2 μm (D, F, and G: inset), 5 μm (A, B, E, I, and J), 10 μm (B, D, F, and G).