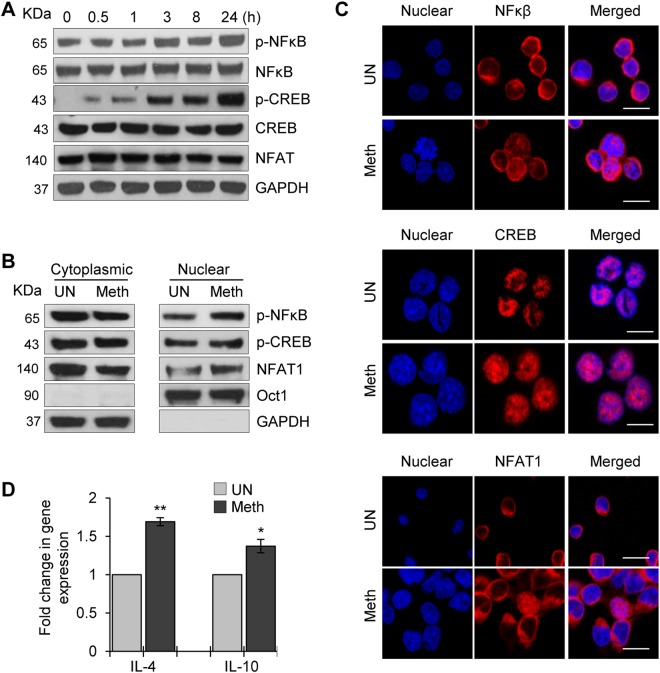
Figure 3.
Meth induced increased phosphorylation and nuclear translocation of CREB, NFAT1 and NFκB in CD4+ T-cells. (A) CD4+ T-cells were untreated or treated with 100 µM Meth for different time points (0.5–24 hours), lysed and the protein extracts were analyzed for the activation of transcription factors NFκB and CREB and NFAT1 by Western blotting. GAPDH was used as a loading control. Full-length blots are presented in Supplementary Fig. S2. (B) CD4+ T-cells were untreated or treated with 100 µM Meth for 1 hour; cytoplasmic and nuclear extracts were isolated and analyzed for activated transcription factors. Oct-1 is nuclear loading control while GAPDH is cytoplasmic control. Full-length blots are presented in Supplementary Fig. S2. (C) Confocal images of NFκB (upper panel), CREB (middle panel) or NFAT1 (lower panel) staining in CD4+ T-cells untreated or treated with 100 µM Meth for 1 hour. Meth treated samples show nuclear translocation of NFκB, CREB and NFAT1. Scale bar = 10 µm. (D) CD4+ T-cells were untreated or treated with 100 µM Meth for 24 hours, RNA was isolated, and IL-4 and IL-10 gene expression was analyzed by qRT-PCR. Fold change was calculated by normalizing the Meth treated cells to untreated cells. Data represent the mean ± SD of 3 independent experiments (*p ≤ 0.05, **p ≤ 0.01).