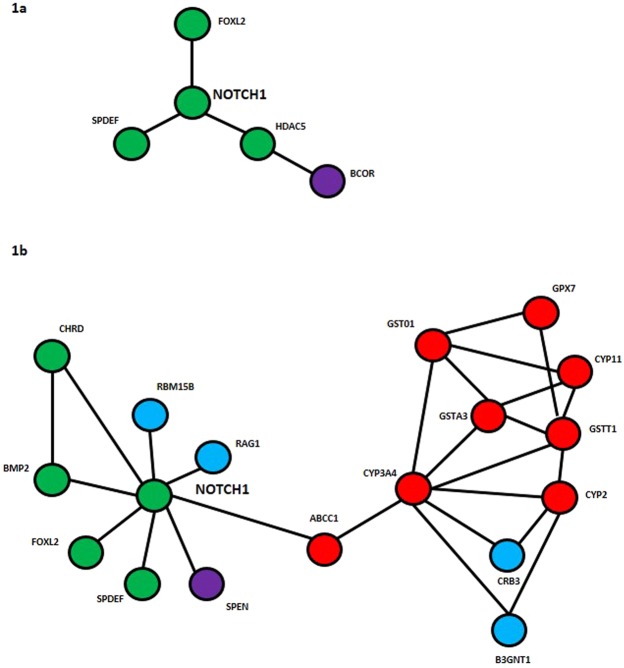
Figure 1.
Schematic diagrams of gene network analyses created in STRING for 3dpf larvae detailing putative protein-protein interactions. Protein functions are colour coded: green – signalling proteins and transcription factors; red – antioxidants and detoxification proteins; purple – transcriptional repressors; blue – varied. (a) Larvae produced from ambient acclimated adults. Gene names: notch1: neurogenic locus notch homolog protein 1; foxl2: Forkhead box L2; hdac5: Histone deacetylase 2; spdef: SAM pointed domain-containing Ets transcription factor; bcor: BCL6 corepressor. (b) Larvae produced from low pH acclimated adults. Gene names, as for (a) plus gpx7, gst01, gsta3, gstt1: members of the Glutathione transferase family; cyp3a4, cyp2, cyp11: members of the CypP450 family; abcc1: Multidrug resistance associated protein 1; crb3: Crumbs protein homolog (involved in morphogenesis); b3gnt1: N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase (transmembrane protein); bmp2: Bone morphogenetic protein; chrd: Chordin precursor; spen: Msx2-interacting protein; rag1: Recombination activating gene (DNA binding protein); rbm15b: RNA binding protein 15B (RNA binding).