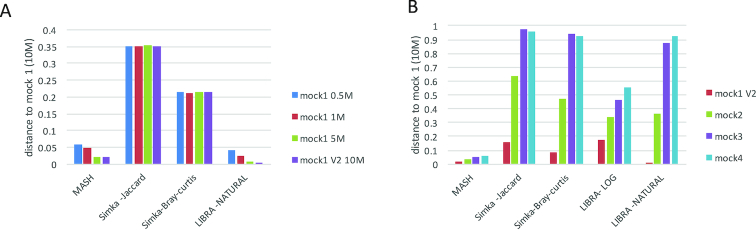
Figure 2:
Analysis of simulated metagenomes using Mash, Simka, and Libra. (A) Distance to staggered mock community simulated metagenome composed of 10 million reads (mock 1 10 M), for simulated metagenomes of same community sequenced at various depth. Simulated metagenomes (454 sequencing) were obtained using GemSim and the known abundance profile of the staggered mock community (see Supplementary Table S2). In order to mimic various sequencing depths, the simulated metagenomes were generated at 0.5, 1, 5, or 10 million reads (noted mock 1 0.5 M; mock 1 1 M; mock 1 5 M; mock 1V2 10 M). The distances between the four simulated metagenomes and a 10 million read simulated metagenome (mock 1 10 M) was computed using Mash, Simka (Jaccard and Bray-Curtis distance), and Libra (natural weighting). (B) Distance to staggered mock community simulated metagenome (mock 1) for simulated metagenomes from increasingly distant communities. The mock 1 relies on the known abundance profile from the staggered mock community. The mock 2 community profile was obtained by randomly inverting three species abundance from mock 1 profile. The mock 3 profile was obtained by randomly inverting two species abundances from mock 2 profile. Finally, a mock 4 profile was obtained by adding high-abundance archeal genomes not present in any the other mock communities. Simulated metagenomes (454 sequencing) were generated using GemSim at 10 million reads. The distance between the mock 1 community to mock 2, mock 3, mock 4, and a replicate community (mock 1 V2) was computed using Mash, Simka (Jaccard and Bray-Curtis distance), and Libra (cosine distance, natural, and logarithmic weighting).