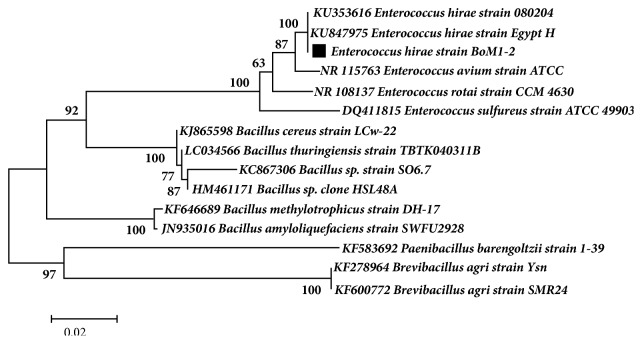
Figure 1.
Phylogenetic tree based on 16S rRNA gene sequences. The evolutionary history was inferred by using the maximum likelihood method based on the Kimura two-parameter model. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Black squares indicate the organisms isolated in this study. GenBank accession numbers of reference sequences are indicated. Bar 0.02 nucleotide substitutions per position.