Figure 1: Overview of the study.
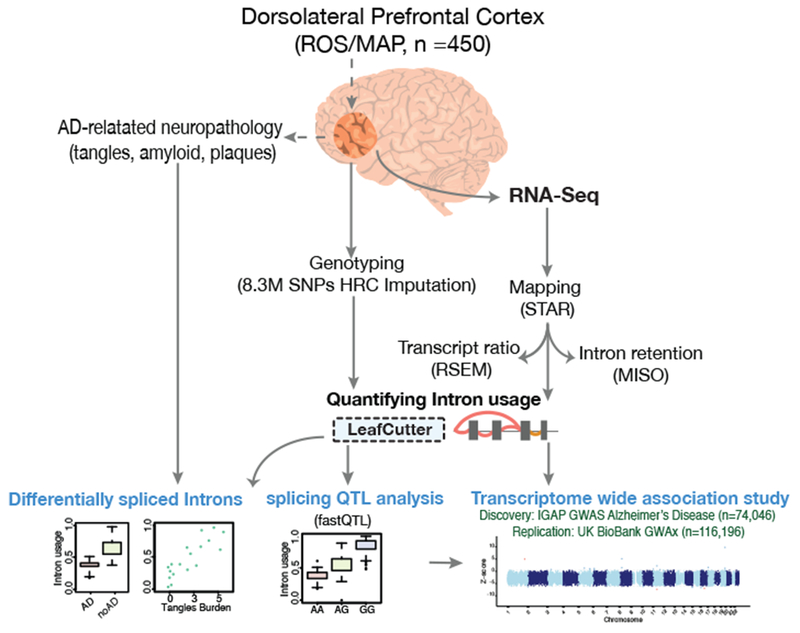
RNA was sequenced from the gray matter of the dorsal lateral prefrontal cortex (DLPFC) of 542 samples (450 remained after QC and matching for genotype data) from the ROS/MAP cohort. RNA-Seq data were processed, aligned and quantified by our parallelized pipeline. The intronic usage ratios for each cluster were then computed using LeafCutter20, standardized (across individuals) and quantile normalized. The intronic usage ratios were used for differential splicing analysis, for calling splicing QTLs, and for transcriptome-wide association studies (TWAS). TWAS was performed on summary statistics from IGAP Alzheimer’s disease GWAS of 74,046 individuals14.
