Figure 6: TWAS prioritizes Alzheimer’s disease genes in endocytosis and autophagy-related pathway.
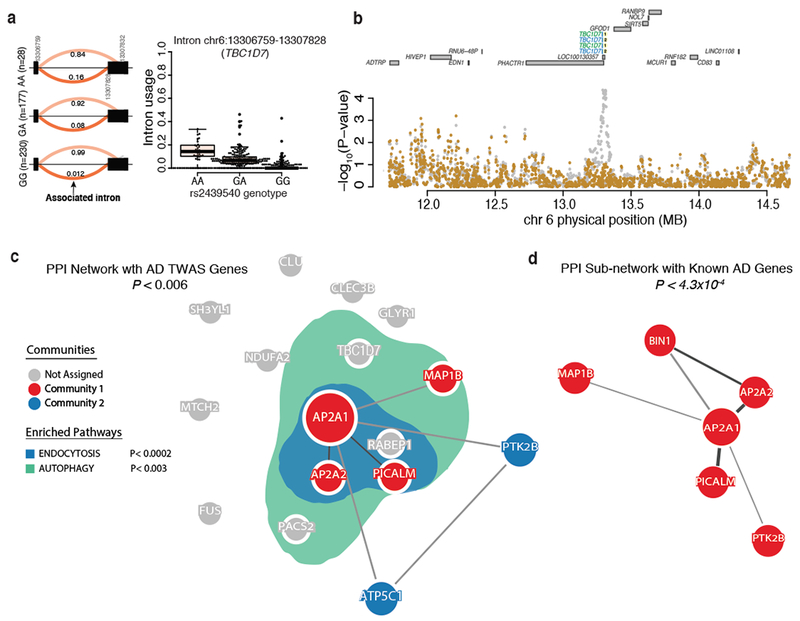
(a) Differential intronic usage for chr6: 13306759:13307828 (TBC1D7) stratified by rs2439540 genotypes (left). Box plot for the same data (right). (b) Regional plot showing the IGAP P-values in TBC1D7 locus. Two intronic excision events at TBC1D7 are present in both ROSMAP (blue) and in CMC (green) dataset. The Alzheimer’s disease GWAS effect is mostly explained by intronic usage of chr6:13306759:13307828. The AD GWAS at TBC1D7 is suggestive in the original IGAP study (p<10−5). (c) The product of three of the novel Alzheimer’s disease genes (AP2A2, AP2A1, and MAP1B) are members of the same PPI network (P < 0.006). The genes in this network and others not in the network (i.e., TBC1D7, PACS2, and RABEP1) are significantly enriched in genes annotated as being involved in endocytosis (blue; P < 0.0002) and autophagy-related pathways (green; P < 0.003). (d) The novel Alzheimer’s disease genes (AP2A2, AP2A1, and MAP1B) form a significant PPI sub-network (P < 4.3 ×10−4) with known Alzheimer’s disease genes (i.e., PICALM, BIN1, and PTK2B).
