Figure 1.
Generation of S2-SLF1 Indel Alleles by CRISPR/Cas9-Mediated Genome Editing.
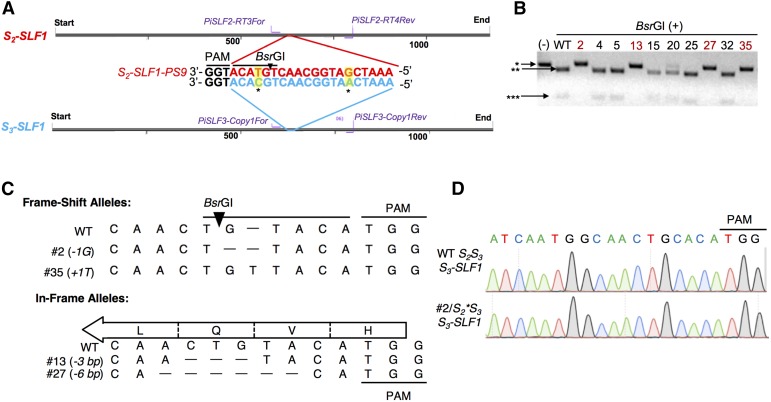
(A) Design of a gRNA specifically targeting S2-SLF1. A 20-bp sequence (named S2-SLF1-PS9) of the antisense strand of S2-SLF1 followed by the PAM motif (TGG) was chosen as the protospacer for CRISPR/Cas9; two mismatches (highlighted in yellow and indicated with asterisks) are found in the corresponding 20-bp region in S3-SLF1 (highlighted in blue). “Start” indicates the start codon (ATG) on the sense strand (5′ to 3′) of these two genes, and “End” indicates the stop codon (TAG). The positions of the PCR primers specific to S2-SLF1 (PiSLF2-RT-3For/PiSLF2-RT-4Rev) and those specific to S3-SLF1 (PiSLF3-Copy1For/PiSLF3-Copy1Rev) are indicated by purple lines. Black triangle indicates the cleavage site of BsrGI in the wild-type S2-SLF1 sequence.
(B) PCR-restriction enzyme digestion screen for edited S2-SLF1 alleles in 10 transgenic plants. (-): PCR product amplified from genomic DNA of one of the transgenic plants by the S2-SLF1 specific primers, without digestion by BsrGI. BsrGI (+): BsrGI digestion of the PCR products amplified from genomic DNA of a wild-type S2S3 plant and the 10 transgenic plants. Asterisk (*) indicates the ∼220-bp PCR product resistant to, or not subjected to, BsrGI digestion; double asterisks (**) indicate the ∼180-bp BsrGI fragment; triple asterisks (***) indicates the ∼40-bp BsrGI fragment. The plant numbers of those T0 plants carrying mutant S2-SLF1 alleles resistant to BsrGI digestion are highlighted in red.
(C) Sequences of four indel alleles in the edited region of S2-SLF1. The sequences shown are those of the antisense strand of S2-SLF1 (5′ to 3′ from left to right). The black triangle indicates the cleavage site of BsrGI in the wild-type S2-SLF1 sequence. The open arrow indicates the direction of translation, and the encoded amino acids in the wild-type S2-SLF1 are shown. The 3-bp in-frame deletion in plant #13 abolishes the codon 5′-CAG-3′ for Gln-210. The 6-bp in-frame deletion in plant #27 abolishes the codon for Gln-210 and disrupts the codon 5′-GTA-3′ for Val-209 and the codon 5′-TTG-3′ for Leu-211. However, as the Val codon is restored as 5′-GTG-3′, only Gln-210 and Leu-211 are deleted from the encoded protein.
(D) Sequencing chromatograms of PCR amplicons of S3-SLF1 from T0 plant #2/S2*S3 and from a wild-type S2S3 plant. The sequences are those of the antisense strand from 5′ to 3′ (left to right).