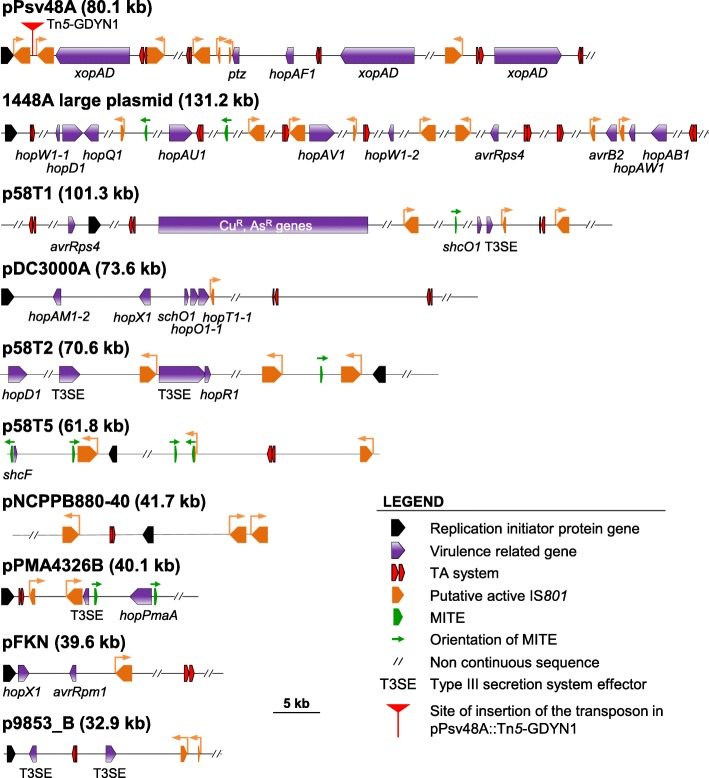
Fig. 6.
Schematic representation of relevant features found in closed plasmid sequences of Pseudomonas syringae. The diagram shows the replication initiator protein genes, virulence genes, TA systems, putative active IS801 elements and MITEs found in closed plasmid sequences of the P. syringae complex. Features are drawn to scale but, for clarity, only pertinent plasmid fragments are shown. The direction of transposition of IS801 fragments and isoforms is indicated with orange arrows. Harbouring organism and accession numbers for the plasmids are P. syringae pv. savastanoi NCPPB 3335, NC_019265 (pPsv48A); P. syringae pv. phaseolicola 1448A, NC_007274 (p1448A); P. syringae pv. tomato DC3000, NC_004633 (pDC3000A); P. cerasi 58T, NZ_LT222313 (p58T1), NZ_LT222314 (p58T2), NZ_LT222317 (p58T5); P. syringae pv. tomato NCPPB 880, NC_019341 (pNCPPB880–40); P. cannabina pv. alisalensis ES4326, NC_005919 (pPMA4326B); P. syringae pv. maculicola M6, NC_002759 (pFKN); P. syringae pv. actinidiae ICMP 9853, NZ_CP018204 (p9853_B)