Fig. 6.
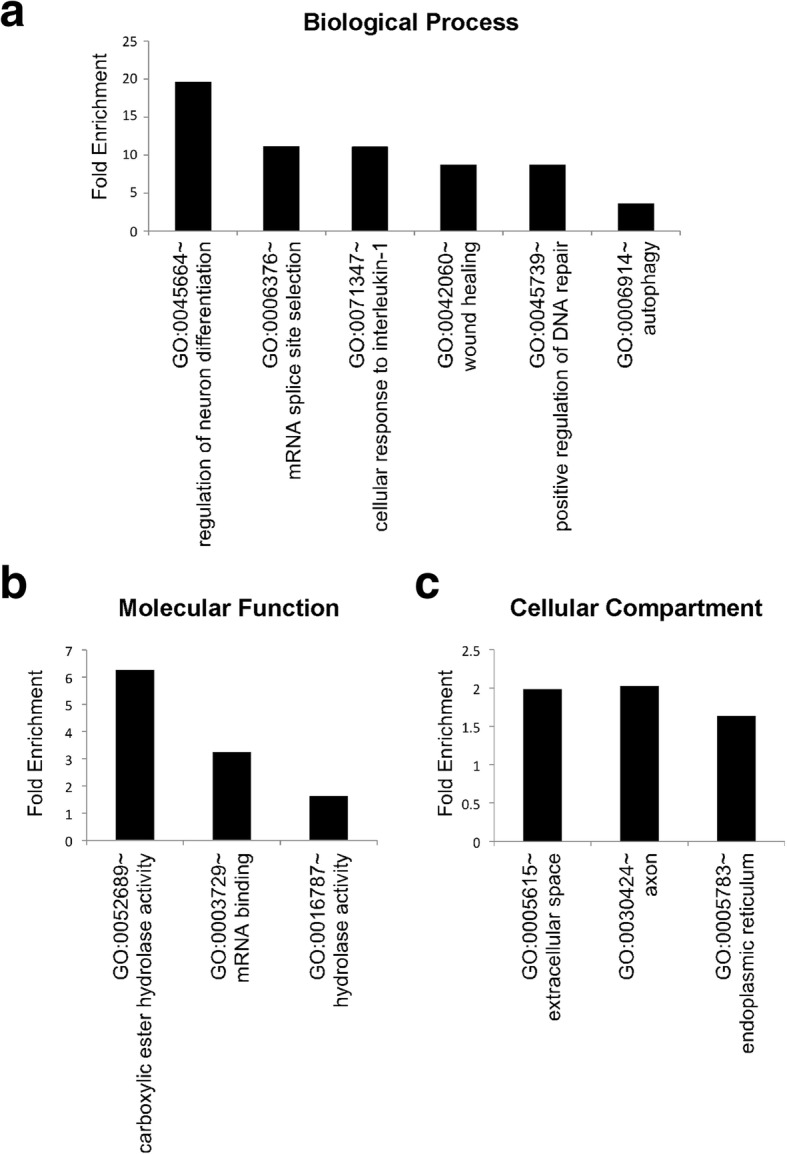
Representation of gene ontology enrichment in mice with constitutive depletion of APMAP, according to biological process (a), molecular function (b) and cellular compartment (c). Label-free quantitative proteomics (procedure described in details in the Materials and Methods section) was used to identify proteins differentially expressed in the brains of APMAP-KO mice. Significantly enriched GO terms (p-value < 0.05) were identified by comparing the list of 113 significantly altered proteins (Additional file 2: Table S1) against the whole list of 2747 detected proteins (Additional file 2: Table S1) using the 1D enrichment tool in Perseus
