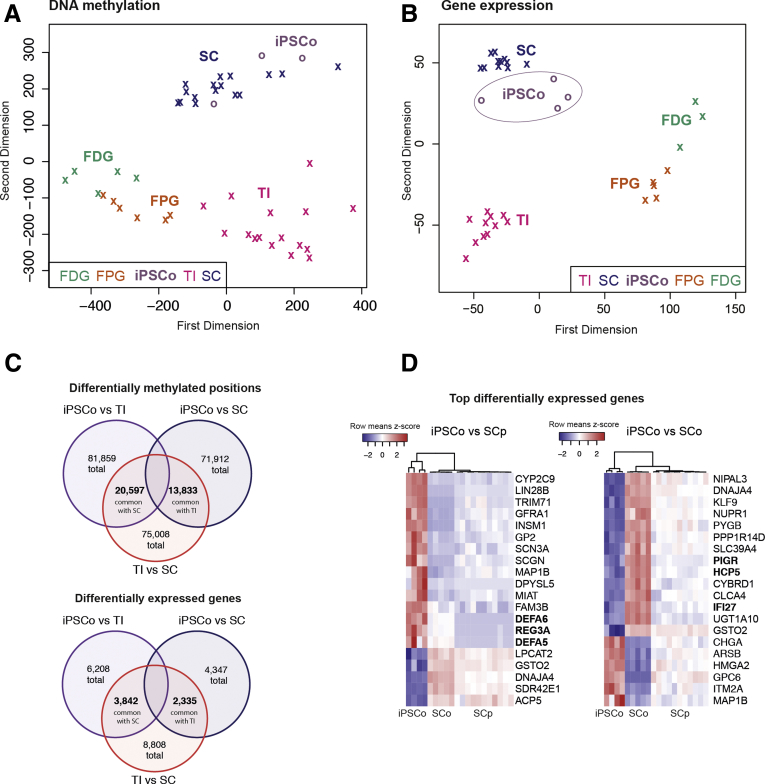
Figure 2.
Genome-wide profiling of human iPSCo and purified IECs. (A) Multidimensional scaling plot showing sample similarity of genome-wide DNA methylation measured by an Illumina 450K array of iPSCo, in the context of IECs derived from terminal ileum (TI), SC, fetal proximal gut (FGP), and fetal distal gut (FDG).1 Each symbol represents a sample, x = IEC, o = organoid. (B) MDS plot of RNA-sequencing profiles showing the same sample groups as in panel A. Sample distance in the plot is based on regularized-logarithmic transformed read counts of all expressed genes. (C) Venn diagram indicating the number of differentially methylated CpG positions (top) or differentially expressed genes (bottom) (adjusted P < .01) between the different groups. (D) Heatmap of expression values (regularized-logarithmic normalized counts) of the top 20 most significantly differentially expressed genes comparing iPSCo and SC purified epithelium (SCp) (left), and iPSCo and SCo (right) for all 3 sample groups. The scale indicates the row mean Z-score (means, 0). The dendrogram above the heatmap clusters samples based on the expression similarity of those genes.