Figure 2.
Deletion of p53 Facilitates Derivation of haiTSCs
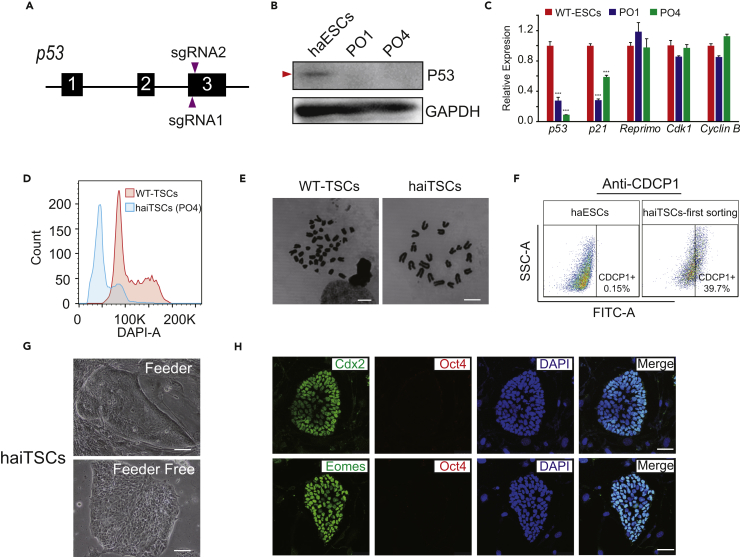
(A) Schematic diagram of the strategy to knock out p53 via the CRISPR/Cas9 system. The two sgRNAs are designed to target exon 3 of p53.
(B) Western blot to detect p53 in PO1, PO4, and WT-haESCs. GAPDH is used as a loading control.
(C) The expression levels of p53-related genes and cell-cycle-related genes (p53, P21, Reprimo, Cdk1, and CyclinB) in PO1, PO4, and WT-ESCs by qPCR. t test, ***p < 0.001. Data are represented as mean ± SEM.
(D) DNA content analysis of haiTSCs derived from the cell line PO4. The percentage of the 1n (G0/G1) peak was 70.4%. Diploid WT-TSCs are used as a control.
(E) Chromosome spreads of haiTSCs and WT-TSCs. haiTSCs have a 20-chromosome set, whereas WT-TSCs show 40 chromosomes in a single cell. Scale bar, 7.5 μm.
(F) TSC-specific CDCP1 antibody analysis of derived haiTSCs at first sorting. The percentage of CDCP1-positive cells is 39.7%.
(G) Images of haiTSCs colonies on feeder cells and on Matrigel. Scale bar, 100 μm.
(H) Immunofluorescence staining of TSC markers (Cdx2 and Eomes, fluorescein isothiocyanate channel) and pluripotent markers (Oct4, Tetramethylrhodamine [TRITC] channel) in haiTSCs. DNA is stained with DAPI. Scale bar, 50 μm.
See also Figure S3.